Abstract
Targeted delivery of anti‐tumor drugs and overcoming drug resistance in malignant tumor cells remain significant clinical challenges. However, there are only few effective methods to address these issues. Extracellular vesicles (EVs), actively secreted by cells, play a crucial role in intercellular information transmission and cargo transportation. Recent studies have demonstrated that engineered EVs can serve as drug delivery carriers and showed promising application prospects. Nevertheless, there is an urgent need for further improvements in the isolation and purification of EVs, surface modification techniques, drug assembly processes, and precise recognition of tumor cells for targeted drug delivery purposes. In this review, we summarize the applications of engineered EVs in cancer treatment and overcoming drug resistance, and current challenges associated with engineered EVs are also discussed. This review aims to provide new insights and potential directions for utilizing engineered EVs as targeted delivery systems for anti‐tumor drugs and overcoming drug resistance in the near future.
Keywords: drug delivery, drug resistance, extracellular vesicles, nanoparticles
Abbreviations
- APCs
antigen‐presenting cells
- Apts
aptamers
- AS1411
a guanine‐rich DNA aptamer
- BBB
blood‐brain barrier
- BC
breast cancer
- CAFs
cancer‐associated fibroblasts
- CP05
specifically recognizes CD63
- DCs
dendritic cells
- DHA
dihydroartemisinin
- dPGs
dendritic polyglycerol
- EGFR
multiple drug resistance
- EGFR
epidermal growth factor receptor.
- EMA
European Medicines Agency
- eNVs‐FAP
FAP gene‐engineered tumor cell‐derived exosome‐like nanovesicles
- EVs
extracellular vesicle
- FAP
fibroblast activating protein‐α
- FAP
fibroblast activating protein‐α
- FDA
Food and Drug Administration
- GRP78
glucose‐regulated protein 78
- HGF
hepatocyte growth factor
- imDCs
immature dendritic cells
- iRGD
rabies virus glycoprotein
- lncRNA
long non‐coding RNA
- MDR
multidrug resistance
- miRNA
microRNA
- mPC
metastatic peritoneal carcinoma
- MSCs
mesenchymal stem cells
- MTTP
microsomal triglyceride transfer protein
- MVB
multivesicular bodies
- ncRNA
non‐coding RNA
- NEs‐Exos
neutrophil‐exosome system
- NK cells
natural killer cells
- NPs
nanoparticles
- PDGFR
platelet‐derived growth factor receptor
- PEG
polyethylene glycol
- PSMA
prostate‐specific membrane antigen
- PSMA
prostate‐specific membrane antigen
- PTK7
protein tyrosine kinase 7
- RNAi
RNA interference
- sdAb
single domain antibody
- siRNA
small interference RNA
- SIRP
signal regulatory protein
- STAT6
signal transducer and activator of transcription 6
- TAMs
tumor‐associated macrophages
- tLyp‐1
truncated LyP‐1
- TME
tumor microenvironment
1. BACKGROUND
The precise targeting of drug delivery and drug resistance pose significant challenges in cancer treatment and are the main causes of treatment failure and detrimental side effects. Chemotherapy, commonly utilized in treatment for tumors at advanced stages, exhibits high toxicity towards normal cells, resulting in a range of adverse reactions, including fatigue, vomiting, rash, intestinal disorders, and febrile neutropenia [1]. Additionally, multidrug resistance (MDR) is a prevalent issue in the use of free (or unbound) chemotherapeutic drugs [2, 3]. Consequently, a pressing need exists to achieve precisely targeted delivery of medications and to combat drug resistance.
In recent years, nanoparticles (NPs) have made considerable strides in drug delivery [4], with a diverse range of nano‐based delivery systems approved by the US Food and Drug Administration (FDA) [5]. Although the development of nanomaterials once gave us hope, the therapeutic efficacy of nanomaterial drugs has not met expectations because of their high expense, poor biological compatibility, and potential to elicit an immune response [6]. Due to their inherent biocompatibility in vivo, extracellular vesicles (EVs) have shown better results in enhancing medication transport and therapeutic benefits than synthetic nanomaterials [7, 8]. EVs, small vesicles varying in size from 30 to 120 nm [8], are secreted by various cell types and can facilitate cell‐to‐cell communication without necessitating direct contact. Therefore, EVs can be utilized as local or long‐range drug delivery systems [9].
EVs possess the advantageous traits of low toxicity, low immunogenicity, and relative stability in vivo, rendering them an attractive option as carriers for nucleic acids and drugs [10, 11]. They can be genetically engineered in their natural state to confer cell and tissue targeting and can safely deliver loaded drugs both in vitro and in vivo [12, 13]. Hence, EVs serve as promising natural nano‐carriers for clinical applications. Various methods exist for the modification, loading, and application of engineered EVs. Here, we summarize the processes used to create modified EVs using genetic or chemical techniques, their use for targeted drug delivery, and their value in preventing drug resistance. Furthermore, the use of engineered EVs for disease treatment through drug packaging as well as insights and prospects for the future of engineered EVs are discussed.
2. NPS FOR TARGET DRUG DELIVERY
The remarkable advancement of nanotechnology in recent decades endowed NPs with the capability to enhance cargo stability and solubility, facilitate transmembrane transport, and extend circulation time to improve safety and efficacy. Consequently, NPs have emerged as a kind of viable medication delivery materials with several benefits [14, 15]. Notably, targeted therapy of NPs predominantly relies on the enhanced permeability and retention effect [16]. Currently, the US FDA and the European Medicines Agency (EMA) have authorized over 25 nanomedicines, with over 45 additional NP technologies under evaluation in clinical trials, despite lacking the US FDA/EMA approval (Figure 1) [17].
FIGURE 1.
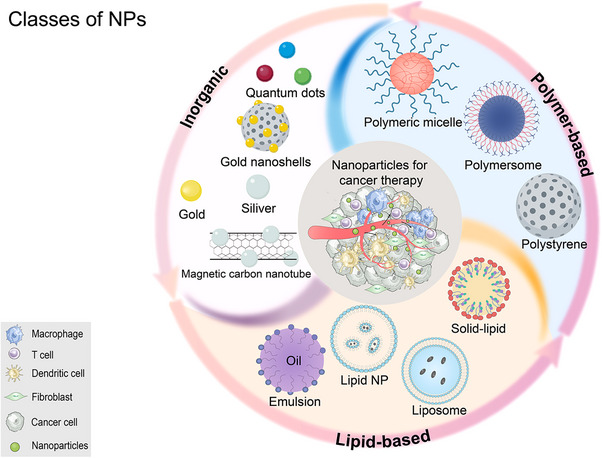
Classes of NPs. Listed here are the three most commonly used types of NPs and their subclasses. NP, nanoparticle
The specific binding between a ligand and a receptor confers the nano‐carrier with active targeting capability [18, 19]. Folate receptors are known to be overexpressed in more than 40% of various carcinomas [20]. As a carrier, folic acid‐modified polymer exhibits high cellular uptake in tumors such as cervical cancer and squamous cell carcinoma and can encapsulate anti‐tumor drugs, including baicalin or nucleic acid, to augment its anti‐tumor effects [21, 22, 23]. The high expression of transferrin in malignant cells and its low expression in normal human tissues render it an appealing molecule for targeted cancer therapy [24]. Graphene oxide was used by Liu et al. [25] as a drug carrier, surface‐modified with transferrin, and loaded with dihydroartemisinin (DHA). Upon injection into mice with breast cancer (BC), the resulting formulation exhibited a remarkable tumor‐targeting ability, responded to a low pH trigger, and significantly enhanced the cytotoxicity of DHA.
In addition to ligand modification, the surface modification of antibodies is another option [26]. Using this feature, some studies have modified graphene quantum dots by attaching a high‐affinity epidermal growth factor receptor (EGFR)‐scFv (B10), enabling precise targeting of BC cells that overexpress EGFR [27, 28]. Upon loading with cisplatin, scFv‐modified NPs effectively killed tumor cells. Similarly, a single domain antibody (sdAb) specific to EGFR was used as a targeted warhead to modify bimodal dendritic polyglycerol (dPGs) that can be cleared through the kidney. The findings indicated that antibody‐modified NPs exhibited enhanced specificity and a greater tendency to accumulate in tumors [26].
Aptamers (Apts) are small, single‐stranded DNA or RNA molecules that have high specificity and affinity but are less immunogenic than monoclonal antibodies [29]. As a result, several nucleic acid Apts have been used to modify NPs to accurately target cancer cells. For instance, to treat prostate cancer, A102‐fluorouracil RNA Apts that target the extracellular domain of prostate‐specific membrane antigen (PSMA) were used to modify NPs and encapsulate docetaxel NP‐Apt. These modifications have demonstrated significant anti‐cancer efficacy and low toxicity in vitro and in vivo [30]. Moreover, A10‐3.2, an RNA Apt, bound PSMA more strongly than A10, and the modified nano‐polymer could inhibit prostate cancer cells with multiple targets and induced selective death by loading miR‐15a and miR‐16‐1 [31]. Interestingly, as for PSMA‐targeting peptides, some researchers constructed natural killer (NK) cells targeting castration‐resistant prostate cancer through lentivirus and showed significant anti‐tumor effects in vivo and in vitro. The anti‐tumor mechanism may be related to iron death [32]. The interesting point of this experiment is that it modifies NK cells directly, which gives full play to the characteristics of NK cells that do not need antigen stimulation and strongly kill tumors. More importantly, compared with nanomaterials, NK cells have higher safety and feasibility [33]. It has great potential in tumor immunotherapy. Thus, despite the numerous advantages of NPs, their actual clinical applications are currently limited due in part to the translational gap between humans and animals [34]. Additionally, there are concerns regarding the toxicity and biological safety of NPs in vivo. When NPs enter the body, they may have an immune reaction with the body (that is, uptake by macrophages), resulting in strong adverse reactions, such as immunosuppression, increased risk of infection, interference with bacterial clearance from the blood, and anaphylaxis [35, 36, 37, 38]. Even when polyethylene glycol (PEG) is used as an “invisibility cloak”, repeated injections may lead to the production of PEG antibodies, resulting in faster removal of NPs [39, 40, 41]. Collectively, NPs are limited by biocompatibility and safety, as well as problems related to large‐scale production, so they are difficult to be widely used in the clinic [42, 43, 44].
Targeted delivery of anti‐tumor drugs and drug resistance for malignant tumor cells are still big challenging clinical issues, but there is still a lack of effective means and methods, including nanomaterials and Apts, to solve them. EVs, a key mediator actively secreted by cells, have been shown to be crucial for intercellular information transmission and cargo transportation [45]. Recently, some studies have shown that engineered EVs can serve as drug delivery carriers and have successfully constructed oral drugs packaged paclitaxel which demonstrating good application prospects [46, 47]. However, further improvements are urgently needed in the separation and purification, surface modification, drug assembly, and precise recognition of tumor cells of EVs as drug‐targeted delivery agents. Thus, in this review, we summarized the current progress of the isolation, purification, surface modification of EVs, as well as their long‐term prospects in targeted delivery of anti‐tumor drugs and overcoming drug resistance for cancers. It will provide new ideas and possible directions for engineered EVs as targeted delivery systems for anti‐tumor drugs and overcoming drug resistance for tumor cells.
3. ENGINEERED EVS AND THEIR THERAPEUTIC POTENTIAL
3.1. EVs
EVs were identified in reticular cells in 1983 and were initially assumed to be responsible for cellular waste removal [48]. However, in‐depth investigations into EVs in recent years have revealed their unique characteristics. EVs are ubiquitously produced and released into the extracellular fluid by almost all cells. They are about 30‐120 nm in size and are considered natural endogenous NPs [49, 50].
EV production starts with the invagination of the cell membrane, which enters the cytoplasm to become early endosomes. These early endosomes receive particulate matter from cells to form late endosomes, which eventually develop into multivesicular bodies (MVB) under endocytosis. An MVB then combines with certain portions of the cell membrane before being released into the extracellular matrix as vesicles (Figure 2) [51, 52]. Since EVs can be released by almost all cells, they possess excellent circulatory stability and have negligible cytotoxicity in vivo [53, 54]. At the same time, EVs can be extracted from patients, so they have low immunogenicity, which has more advantages than NPs [55, 56, 57]. EVs offer distinct benefits in treating brain diseases owing to their capacity to quickly penetrate the blood‐brain barrier (BBB), according to recent research [58]. Compared with directly delivering live cells, EVs have shown encouraging results in the treatment of brain diseases [59, 60].
FIGURE 2.
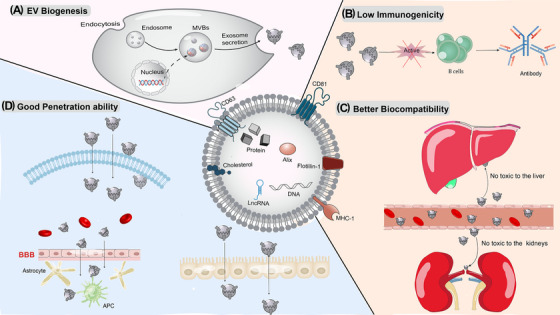
The formation and biological characteristics of EVs. (A) The formation of EVs; (B) Low immunogenicity. (C) Only a small number of EVs will accumulate in places such as the liver and kidneys, so there is little toxicity in the body; (D) EVs can pass through many kinds of biological barriers, such as the plasma membrane, the BBB and the cell barrier. APC, antigen‐presenting cell; BBB, the blood‐brain barrier; EV, extracellular vesicle; lncRNA, long non‐coding RNA; MHC, major histocompatibility complex; MVB, multivesicularbody
3.2. Characterization of EVs
As for how to identify EVs, the International Society for Extracellular Vesicles proposes that at least one of the following proteins needs to be evaluated (e.g., the plasma membrane‐associated or endosome‐associated transmembrane or glycosylphosphatidylinositol‐anchored extracellular proteins and lipid‐ or membrane protein‐binding cytoplasmic proteins) to determine whether they are EVs [61]. In most studies, electric vehicles are typically characterized using three methods: transmission electron microscopy, the particle diameter analysis system and western blotting [61, 62, 63]. EV transmission electron microscopy can identify the size and shape of EVs to determine the presence of an EV‐like structure in the sample, which is usually in a saucer shape with a concave center or a hemispherical shape with a concave side [64]. The original state of EVs can be ensured by measuring and counting the diameter distribution of particles in the sample through the particle diameter analysis system. Western blotting identification of EV surface protein markers: usually the detection of protein markers, CD9, CD63, and tumor susceptibility 101 (TSG101), is used to determine the presence of EVs. In addition to the above three general methods, EV markers can also be detected by flow cytometry, which has the advantages of fast analysis and fewer samples required. However, this technology is time‐consuming and labor‐intensive, which limits its applications [65]. In addition, a time‐resolved fluorescence immunoassay based on NPs has been developed, which can capture EVs directly from urine and cell supernatant and identify disease‐specific markers on the surface of urine EVs from patients [66]. Therefore, researchers can actively select and develop a variety of markers to characterize EVs in many ways according to their own needs and conditions.
3.3. Isolation of EVs
There are many methods for isolating EVs, and different separation methods can be used according to different purposes and needs. Among them, ultracentrifugation, ultrafiltration, precipitation, immunoaffinity capture, and size exclusion chromatography are the five most commonly used techniques, and these techniques may be used in combinations in different ways.
Ultracentrifugation is the gold standard for the isolation of EVs and is the most commonly used isolation technique. This method separates components in a solution based on differences in size and density [67], and the operation is simple. In addition to the initial instrument, the price is low, and a high concentration of EVs can be separated [68]. However, its disadvantages are time‐consuming, high equipment requirements, and damage to the structure [69].
Ultrafiltration is a selective separation based on the size of particles. This method is faster than ultracentrifugation, and the activity of EVs is not affected. However, there are problems, such as damage to EVs due to the applied shear stress and low purity of recovery [70].
The precipitation method uses PEG to harvest EVs by reducing the solubility of EVs [71]. This method is relatively simple to operate and is used to separate large‐dose samples. However, the purity and recovery rate of EVs are low, which is not conducive to subsequent functional experimental analysis [72].
Immunoaffinity capture is based on the fact that EVs can be separated by recognizing their unique surface markers (such as CD9, CD63, CD81 and Alix) through antibodies immobilized on surfaces such as magnetic beads, chromatography column resins, multi‐well plates and microfluidic device [73]. This method has the characteristics of strong specificity, high sensitivity and high purity [74]. However, there are also some problems such as high cost and unsuitability for large‐scale separation [75].
Size exclusion chromatography, the mildest chromatographic technique, takes advantage of size differences, that is, the differential elution of particles of different sizes through gravity flow, thus effectively separating EVs from most proteins (such as albumin) in body fluids. This method can maintain the integrity and biological activity of EVs and has high yields, but it is time‐consuming and has a low recovery rate [76].
In general, each method has advantages and disadvantages, and researchers need to integrate and innovate existing methods according to their own needs. For example, to improve the separation purity, some studies have tried to combine ultracentrifugation and ultrafiltration with size exclusion chromatography to isolate EVs [77, 78]. The results showed that ultrafiltration combined with size exclusion chromatography could reduce the level of impurity cytokine (interleukin‐10) in isolated EVs.
3.4. Therapeutic potential of EVs
The secreted EVs can engage with corresponding target cells through a diverse range of mechanisms [79]. Upon receptor binding, it initiates fusion and endocytosis processes, subsequently triggering relevant biological responses [80, 81]. Nucleic acids, including microRNA (miRNA), long non‐coding RNA (lncRNA) and DNA, may be transported via EVs to facilitate cell‐to‐cell communication. According to Liu et al. [82], EVs from activated T cells contain miRNAs that regulate immunity by targeting antigen‐presenting cells (APCs). Similarly, EVs produced by donor dendritic cells (DCs) have been shown to include functional donor major histocompatibility complex molecules and APC activation signals, which may increase allograft rejection [82]. Moreover, EVs can convey proteins expressed by parent cells to facilitate intercellular communication. For example, EVs can transfer a variety of proteins to promote the development of illnesses, including Alzheimer's disease and prions [83, 84].
These observations highlight the natural EVs' effectiveness as a drug delivery platform in light of their biodistribution, biocompatibility, and minimal immunogenicity. EVs have a high permeability and are capable of traversing most biofilms, including the BBB (Figure 2) [57]. Moreover, as they come from the patients’ own body, the likelihood of immune response, allergic reaction, toxicity, and biodegradability issues is low [85]. Given these advantages, EVs hold promise as candidates for cancer therapeutic delivery.
4. SURFACE MODIFICATION OF EVS
Like other nano‐therapeutic drugs, natural EVs in animals tend to amass in organs such as the liver, kidney, and spleen upon systemic delivery and are rapidly cleared by phagocytosis in the reticuloendothelial system [86]. Therefore, their surfaces require specific modification to achieve accurate targeting to specific cells or tissues, thereby mitigating their accumulation in healthy tissues. Surface engineering's primary goal is to impart targeted specificity to certain cell types. Genetic engineering and chemical alteration are examples of modification approaches (Figure 3).
FIGURE 3.
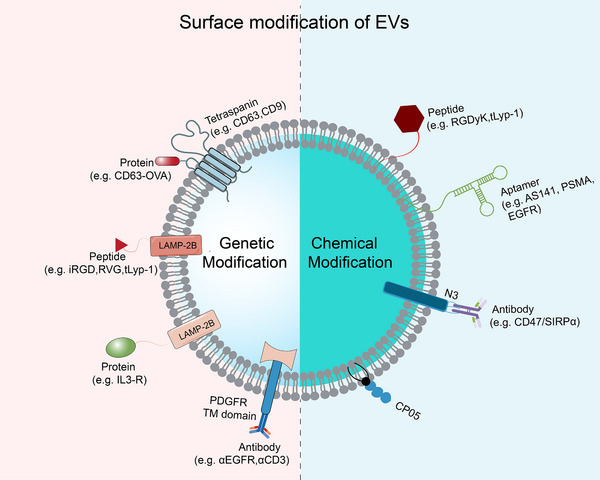
Alteration of EV surface by chemical and genetic engineering. Chemical lipid or membrane‐bound protein processes or lipid‐lipid interactions load many components, such as peptides, nucleic Apts and antibodies. Through the joining of membrane‐binding proteins, genetic engineering introduces targeted sequences, including peptides and proteins. Apts, aptamers; AS1411, a guanine‐rich DNA aptamer; CP50, the polypeptide fragment CP05 specifically recognizes CD63; EGFR, epidermal growth factor receptor.CD63‐OVA, an ovalbumin antigen fused with CD63; EV, extracellular vesicle; PDGFR TM domain, the transmembrane domain of platelet‐derived growth factor receptor; PSMA, prostate‐specific membrane antigen; RVG, rabies virus glycoprotein; tLyp‐1, truncated LyP‐1
4.1. Chemical modification
Chemical modification of EVs can improve their physical stability and increase the effectiveness of both their targeting and medication delivery [87]. One effective method for co‐binding small and macromolecular organisms to the EVs’ surface is click chemistry, which has a quick reaction time, high specificity, and aqueous compatibility. In addition, click chemistry has minimal effects on the function of EVs [88]. To address the challenges of treating gliomas, in which the BBB often restricts the entry of drugs into the brain, thus affecting the treatment of tumors, Jia et al. [89] coupled the EVs’ membrane with the neuroderm‐1 targeting peptide by click chemistry to deliver curcumin and NPs using EVs. The resulting EVs could pass through the BBB and treat gliomas simultaneously. Similarly, c(RGDyK) peptide, which demonstrated a strong affinity for integrin α(v)β3 in reactive cerebral vascular endothelial cells after ischemia [90, 91], was found attached to the EVs’ surface through a chemical reaction, and the curcumin‐loaded c peptide was then intravenously delivered into a mouse model of transient middle cerebral artery blockage [59]. The c peptide EVs can effectively penetrate the BBB and inhibit inflammation and apoptosis in the lesion area [59].
Chemical modification can also be used to inhibit tumor immune escape. “Don't eat me” signals are activated when CD47 binds to the signal regulatory protein (SIRP) on innate immune cells, including macrophages and DCs, which allow the tumor to evade phagocytosis [92, 93]. By using click chemistry via a pH‐sensitive junction, Nie et al. [94] coupled an M1 macrophage‐modified EVs with dibenzocyclooctyne‐modified antibody of CD47 and SIRP. After systemic administration, the modified EVs recognize active tumor targeting and effectively reprogram into an M1‐dominated tumor microenvironment (TME).
Nucleic Apts are a promising tool for specific targeting owing to their strong affinities and selectivity for target molecules [95]. The EVs' surface can be modified with nucleic Apts to improve their ability to target cells and increase the effectiveness of medication delivery [96, 97]. For instance, Apts (AS1411) that target nucleolin, a protein that is overexpressed on the surface of BC cells, were added to EVs. These AS1411‐modified EVs efficiently delivered small interference RNA (siRNA)/miRNA to BC cells in mice, resulting in effective tumor inhibition [98]. Another study used an Apt (sgc7) that recognizes protein tyrosine kinase 7 (PTK7) to modify the surface of EVs for targeted delivery of chemotherapeutic drugs to cancer cells expressing PTK7 [87]. Additionally, EVs have been modified with RNA Apts (such as PSMA‐Apts and EGFR‐Apts) to efficiently deliver siRNA/miRNA to corresponding tumor sites, leading to substantial tumor growth suppression. Overall, EV nucleic Apt modification has the potential to significantly increase the specificity and effectiveness of targeted medication delivery [99].
The EVs’ surface can also be modified by other chemical methods. Qi et al. [100] created a cluster of superparamagnetic NPs based on EVs with dual functions that connect transferrin‐bound superparamagnetic NPs to the EVs derived from blood reticulocytes by transferrin‐transferrin interaction, which not only enhances the specificity and efficiency of cancer targeting but also enables real‐time imaging and monitoring of the drug delivery process. Nakase et al. [101] combined cationic lipid treatment with the negatively charged membrane of EVs and added lipodiamine to the EVs’ surface, which greatly improved the cell uptake efficiency. CP05 anchoring modification has been proven to retain the natural size and morphological characteristics of EVs without affecting their distribution in vivo and can change their cell targeting behavior, thus more accurately treating tumors [102].
However, the availability of certain functional groups on the surface of the EVs may restrict chemical modification, and it may not be possible to target certain molecules using this approach [103]. Furthermore, it is crucial to properly account for any possible safety concerns brought on by chemical alterations, especially when designing EV‐based therapies for clinical use. The modified EVs must not be harmful and must not cause the patient to develop an immunological reaction. Additionally, regulatory hurdles may need to be addressed before modified EVs can be approved for clinical use.
4.2. Genetic engineering
Compared to chemical modification, genetically engineered EVs offer several advantages for the expression and stability of the targeted parts [104]. Moreover, the EV membrane's diversity of transmembrane proteins, including lysosomal‐associated membrane protein (LAMP) and glycosylphosphatidylinositol, can be fused with ligands or homing peptides to enhance the transport of EVs to their intended destination [105].
By using genetic engineering, the gene sequences for the relevant surface membrane protein and the ligand or homing peptide may be combined. A common target for the genetic engineering of EVs for targeted distribution is LAMP‐2B, a transmembrane protein that is abundant in EVs generated from DCs [106]. In this approach, the targeting protein or peptide's gene is fused with LAMP‐2B's gene to generate a chimeric protein that is produced on the EVs’ surface. Despite this limitation, targeted medication delivery using genetically modified EVs has shown encouraging results, and the potential for therapeutic use is now being extensively investigated. Kumar et al. [107] used EVs derived from DCs to fuse the EVs’ membrane protein LAMP‐2B into the central nervous system specific rabies virus glycoprotein (RVG) peptide. The RVG peptide can specifically bind to neuronal cells to express acetylcholine receptors. In mice, glyceraldehyde‐3‐phosphate dehydrogenase siRNA‐loaded RVG‐targeting EVs were injected intravenously. The results demonstrated that EVs for RVG targeting might selectively target brain neurons and result in specific gene silencing. This provides a promising vector for the treatment of glioblastoma and gene therapy of chronic neurodegenerative diseases [108]. Given that immature dendritic cells (imDCs) can generate EVs with low immunogenicity and toxicity [109, 110], Tian et al. [111] employed imDCs and fused the N‐terminal of the mouse EV LAMP‐2B protein with the rabies virus glycoprotein (iRGD) peptide, which is specific for integrin α(v)β3. Experiments revealed that iRGD EVs could specifically deliver adriamycin or doxorubicin to integrin‐positive BC cells. Bai et al. [112] fused the targeted peptide tLyp‐1, a ligand of selective targeting neuropilin 1 and neuropilin 2 [113, 114], to LAMP‐2B and successfully constructed targeted tLyp‐1 EVs, which have a high transfection efficiency for use in gene therapy. Given that endosomal proteases may destroy the peptides on the surface of EVs during their synthesis, Hung et al. [115] used genetic engineering methods to combine the N‐terminus of the targeting peptide‐LAMP‐2B fusion with the glycosylation sequence to prevent peptide degradation.
LAMP‐2B is not the only transmembrane protein that can be used for membrane display. Another option is to use platelet‐derived growth factor receptor (PDGFR), a transmembrane protein. Cheng et al. [116] combined antibodies targeting T cell CD3 and cancer cell‐associated EGFR with the transmembrane domain of human PDGFR to create EVs that showed powerful anti‐cancer immunity against EGFR‐positive triple‐negative BC cells in vitro and in vivo.
In addition to PDGFR and LAMP‐2B, tetraspanin superfamily proteins CD63, CD9, and CD81 can be designed to display targeting sequences or probes [117, 118, 119].
However, genetic engineering‐based modification is limited by the encoded gene sequence, and its application is challenging. Ongoing researches are focusing on improving this method. In summary, surface modification of EVs significantly enhances their stability and targeting, but it is a challenging process that requires strict control of reaction conditions to prevent destruction and non‐specific accumulation of EVs due to inappropriate temperature and pressure [120].
5. THERAPEUTIC CARGO LOADING IN ENGINEERED EVS
EVs can carry various types of cargo, the most common of which are small molecular drugs and nucleic acids (Table 1). According to the loading mode, it can be divided into endogenous loading and exogenous loading (Figure 4). Endogenous loading is through the transformation of donor cells to secrete EVs that we are interested in, which can be achieved by transfection or co‐incubation. Co‐incubation is suitable for some fat‐soluble drugs, such as curcumin and adriamycin, and nucleic acid molecules, such as siRNA. When co‐incubated, cells can absorb goods by endocytosis and secrete EVs of interest [121, 122]. In the transfection method, a specific plasmid can be transfected into the cell to express the desired goods (such as proteins, peptides, nucleic acids and other active molecules), and then be encapsulated and secreted by EVs. This method is simple to operate, but the loading efficiency is low, and the transfection reagent is toxic to the cell [123, 124]. Exogenous loading is to first purify EVs and then load the goods by physical or chemical methods, including electroporation, ultrasonic treatment and extrusion [125], in which proteins, CRISPR‐Cas9 systems and DNA can be electroporated with high encapsulation efficiency but require purification and may damage the integrity of the EVs’ membrane. The chemical load mainly includes saponins permeation and calcium chloride, in which the use of saponins auxiliary method can greatly increase the drug loading [126]. Interestingly, a recent study has used lipid NPs to deliver functional mRNAs to cells, such as vascular endothelial growth factor‐A (VEGF‐A) mRNA, which can be transformed into EVs secreted by donor cells, and these EVs can effectively induce VEGF‐A‐dependent angiogenesis [127]. Compared with traditional transfection, this method exerts less toxicity to donor cells and has a certain loading efficiency. Therefore, the way of loading is not the same. It is hoped that more researchers will creatively develop more and more effective loading methods in the future.
TABLE 1.
The application of EVs as cargo delivery systems
| Source of EVs | Therapeutic cargo | Disease | Reference |
|---|---|---|---|
| Subcutaneous fat MSCs | miRNA‐199‐3p | Osteoarthritis | [196] |
| HEK293T cells | Interleukin‐10 | Inflammatory bowel disease | [197] |
| Normal human foreskin fibroblast cells | siRNA and shRNA | Pancreatic cancer | [198] |
| HEK293T cells | 5‐FU and miR‐21 | Colorectal carcinoma | [191] |
| Macrophage cells | Paclitaxel | Lung cancer | [169] |
| Raw 264.7 macrophage cells | Doxorubicin and paclitaxel | Multidrug resistant cancers | [148] |
| Hepatic stellate cells | Cas9 RNP | Liver diseases | [62] |
| Human mesenchymal stem cells | MicroRNA‐29a‐3p | Glioma | [199] |
| Primary dendritic cells | VEGF siRNA | Breast cancer | [98] |
| HEK293T cells | ASO‐STAT6 | Colorectal cancer | [160] |
| Milk | Paclitaxel | Lung cancer | [184] |
| H22 hepatocarcinoma cells | Doxorubicin and 5‐FU | Hepatocarcinoma | [200] |
| Raw 264.7 macrophages | Doxorubicin | Colon cancer | [201] |
|
KPC689 cells MSCs |
CRISPR/Cas9 | Pancreatic cancer | [202] |
| [object HTMLSpanElement]MSCs [object HTMLSpanElement]MSCs | siRNA | Huntington's disease | [203] |
Abbreviations: 5‐FU, 5‐fluorouracil; ASO‐STAT6, a signal transducer and activator of transcription 6 targeting antisense oligonucleotides;HEK293T cells, human embryonic kidney 293T cells; miRNA, microRNA; MSCs, mesenchymal cells; RNP, ribonucleoprotein; shRNA, short hairpin RNA.; siRNA, small/short interfering RNA; VEGF, vascular endothelial growth factor.
FIGURE 4.

Therapeutic cargo loading methods for EVs. There are two main ways to load EVs, endogenous loading and exogenous loading, and different loading methods have their own advantages and disadvantages. EV, extracellular vesicle; lncRNA, long non‐coding RNA; miRNA, microRNA
5.1. Delivery of nucleic acids
The objective of gene therapy is to cure diseases brought on by faults and aberrations of genes without causing harmful side effects by introducing foreign, normal genes into target cells [128]. Various diseases, including cancer, can be treated with gene therapy using non‐coding RNA (ncRNA) [129, 130]. miRNA and siRNA can specifically inhibit the expression of any cancer‐related genes/mRNA through RNA interference (RNAi) [131], such as Bcl‐2 [132], c‐Myc [133], KRAS [132, 134], etc. However, ncRNA is a macromolecule that is difficult to deliver within the body. Excitingly, a study demonstrated that the EVs secreted by THP‐143 cells transfected with miR‐1 mimics in vitro can successfully load miR‐143 and can be targeted to tumor cells in mice [135]. Given the absence of delivery vectors that can successfully traverse the BBB, Alvarez‐Erviti et al. [108] began to study targeting engineered EVs to the brain. They skillfully used DCs successfully transfected with LAMP‐2B‐modified pEGFP‐C1 to secrete EVs expressing RVG, which were then electroporated with siRNA and administered to mice. A strong RNAi response was observed throughout the brain: the damaging β‐amyloid 1‐42 protein in the animal cerebral cortex was decreased by 55%, and the mRNA of the beta‐site APP‐cleaving enzyme 1, a protein implicated in the formation of Alzheimer's disease [136], was knocked down by 61%. Surprisingly, only a slight RNAi effect was detected in the liver and spleen. Importantly, there was almost no toxicity and very low immune response after systemic administration [108]. The efficacy of modified RVG in siRNA distribution was further supported by research employing hepatocyte growth factor (HGF) siRNA‐loaded EVs from human embryonic kidney cells [137].
In comparison to siRNA, miRNA can cause target mRNA degradation or translation inhibition without completely binding to its target mRNA sequence. Nie et al. [138] exploited the EVs derived from BC cells that overexpressed integrin β4, as it could specifically act on surfactant C and be specifically internalized by cancer cells. When miRNA‐126 was loaded, the PTEN/PI3K/AKT signaling pathway was blocked, and the growth and invasiveness ability of A3 lung cancer cells were significantly inhibited [138]. More specifically, miRNA's anticancer capabilities may be strengthened by EVs. As demonstrated by the Kuroda laboratory, engineered EVs loaded with miRNA are precisely targeted to BC cells overexpressing EGFR through peptide‐targeting ligands, resulting in reduced growth of cancer cells [139]. Although still in the early stages, these studies together demonstrate the potential of engineered EVs in nucleic acid delivery therapy.
5.2. Drug delivery
One important argument for using engineered EVs as medication delivery carriers is their capacity to overcome drug restrictions and lessen toxicity in the human body. Additionally, EVs are a biological source, and clinical studies have shown that their immunogenicity is minimal [140, 141]. By loading drugs into EVs, we may not only reduce the limitation due to chemical properties of some drugs, such as hydrophobicity, but also improve the circulation time and efficacy in vivo and even reverse drug resistance in tumors. Curcumin, a phenolic compound with anti‐inflammatory, anti‐tumor, antioxidant, and chemopreventive activities [142, 143], has poor clinical availability due to its hydrophobicity, fast metabolism, and rapid systemic elimination [144]. However, the co‐incubation of curcumin with EVs and subsequent delivery throughout the body has been demonstrated to retain the therapeutic activity of the drug, improve its solubility, increase its circulation time in vivo, and enhance its overall utilization [145]. A recent study suggested that loading doxorubicin into EVs secreted by mesenchymal stem cells (MSC‐EXO) may be a superior option due to the excellent low immunogenicity, multi‐directional differentiation, and homing ability of MSC‐EXO [146]. Electroporation of doxorubicin into MSC‐EXO not only reduces systemic dosing but also increases cytotoxicity, inhibits tumor growth [111, 147], and even overcomes drug resistance [148]. Additionally, compared to free drugs, drugs with engineered EVs demonstrated better internalization and longer circulation ability [149].
Aside from intravenous injection, oral administration is more convenient and suitable for patients. EV secretion in milk has been proven to be stable under the conditions of strong gastric acid and intestinal degradation, making it a more effective carrier for oral administration than free drugs [150]. Research has also found that drugs loaded with engineered EVs derived from milk have lower half maximal inhibitory concentration and do not cause immune response or inflammation in vivo, demonstrating better efficacy than free drugs [151].
6. APPLICATION OF ENGINEERED EVS IN THE TREATMENT OF DISEASES
6.1. Application of engineered EVs in tumor therapy
The formation of a tumor involves multiple factors and is a complex process with many characteristics, such as unlimited proliferation, immune escape, resistance to apoptosis, and gene mutation [152]. It needs to have a complete operating system against the body, and every factor that is not conducive to its growth will be suppressed. For example, missense mutations of the p53 gene occur in almost all progressive tumors, thus downregulating its function [153]. This is the so‐called crackdown on dissidents and wooing partners. According to the “barrel principle,” as long as we attack and destroy the original balance of the tumor from one aspect, the growth of the tumor can be inhibited, which gives rise to a variety of treatment methods, such as immunotherapy for TME immunosuppression, RNAi directly acting on the molecular mechanism, and so on [154]. These drugs also face pharmacokinetic challenges in terms of poor targeting specificity and systemic circulation, cell uptake and endosomal escape. Engineered EVs can solve these problems because of their good biocompatibility, strong inclusiveness, and low immunogenicity (Figure 5).
FIGURE 5.
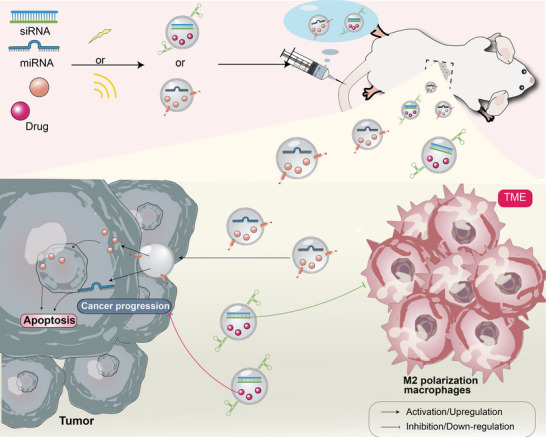
Engineered EVs for tumor treatment. Nucleic acids (such as miRNA) and/or drugs can be loaded into modified EVs by endogenous loading and exogenous loading, such as electroporation or co‐incubation, and then injected intravenously into mice. Such engineered EVs not only reshape TME (such as inhibiting M2 macrophage polarization) but also directly mediate tumor apoptosis or inhibit tumor progression. miRNA, microRNA; siRNA, small/short interfering RNA; TME, tumor microenvironment
6.1.1. Targeting the TME
The TME has a significant impact on patient prognosis, as it greatly affects the effectiveness of chemotherapy [155]. Engineered EVs have been used to alter the TME, facilitating drug accumulation and penetration, improving drug efficacy, and making tumors more responsive to specific therapies. Hu et al. [156] designed a tumor vaccine, FAP gene‐engineered tumor cell‐derived exosome‐like nanovesicles (eNVs‐FAP), which targets cancer‐associated fibroblasts (CAFs). They achieved this by genetically engineering EV‐like vesicles derived from tumor cells to fuse with fibroblast activating protein‐α (FAP). Upon systemic injection in mice, eNVs‐FAP demonstrated a robust anti‐tumor effect, as they not only enhanced anti‐tumor immune cell infiltration but also significantly inhibited the recruitment of immunosuppressive cells, thus remodeling the TME. Moreover, eNVs‐FAP can suppress tumor development and induce ferroptosis [156]. Type M2 tumor‐associated macrophages (TAMs) exhibit tumor‐promoting and immunosuppressive properties in the TME and are often linked to poor outcomes [157, 158]. Type M1 TAMs are mainly pro‐inflammatory and anti‐tumor [159]. To reprogram the TME to the pro‐inflammatory M1 phenotype, Kamerkar et al. [160] used engineered EVs loaded with antisense oligonucleotides of signal transducer and activator of transcription 6 (STAT6) to selectively silence the expression of STAT6 (a key regulator of M2 macrophages [161]) in TAMs, resulting in more than 90% inhibition of tumor growth.
Tumor cells also evade immunity by creating hypoxia and changing the pH of the TME [162, 163, 164]. To address this challenge, a recent study has developed engineered EVs that can respond to changes in pH, allowing the antibodies they carry to be released in the acidic TME, thus effectively reprogramming the TME [94]. Furthermore, engineered EVs that are responsive to high levels of glutathione expression in tumors can be biodegraded locally, releasing catalase, which produces oxygen to alleviate tumor hypoxia and enhance sonodynamic therapy for glioblastoma [165]. These results demonstrated that engineered EVs can improve the microenvironment by inhibiting or reprogramming tumor‐related cells and reprogram the TME by improving environmental factors, thus affecting tumor growth. According to multiple studies, tumor cells can promote tumor growth and immune escape by producing metabolites [166]. These observations raise the question of whether they improve their energy metabolism and inhibit their growth by engineering EVs. Moreover, can the senescent cells [167] and microbiota of similar tumors [168] also be the direction of engineering EVs? These questions need further investigations.
6.1.2. Targeting tumor cells
Tumor cells possess the ability of indefinite self‐renewal and genetic variation, which confer a robust resistance to treatment. However, the advent of engineered EVs provides a glimmer of hope. These EVs can precisely transport medication to the tumor location. Engineered EVs have been developed to identify sigma receptors overexpressed in lung cancer cells [169]. In a mouse model of lung metastasis, its EVs loaded with paclitaxel had superior anti‐tumor effects than free paclitaxel, and the animals' survival time was extended [169]. Metastatic peritoneal carcinoma (mPC) is a common evolution of digestive tract cancer for which there is currently no effective treatments [170]. To explore an effective treatment, Lv et al. [171] synthesized EV‐thermosensitive liposome hybrid NPs through genetic engineering and combined the co‐administration of docetaxel with granulocyte‐macrophage colony‐stimulating factor. The study demonstrated that mPC development was effectively prevented. Drug‐loaded exosomes can also target inflammatory regions. Wang et al. [172] developed a neutrophil‐exosome system (NEs‐Exos), which not only quickly crosses the BBB but also reacts to inflammatory triggers and hunts down tumor cells that are present in inflammatory areas. NEs‐Exos loaded with doxorubicin were intravenously administered into a glioma mouse model. The results demonstrated that NEs‐Exos effectively inhibited the growth of tumor cells and prolonged the survival time of the mice.
For the treatment of tumor cells, the engineered EVs carrying drugs, miRNA, or siRNA can inhibit specific oncogene expression or increase tumor suppressor gene expression, and then they may be used as a target in terms of apoptosis and telomere inhibition and so on.
6.2. Applications in other diseases
Numerous liver diseases, encompassing acute liver injury and chronic liver fibrosis, currently lack satisfactory treatment options, necessitating the search for alternative therapeutic interventions. In comparison to synthetic non‐viral delivery vectors, vectors naturally present in the body possess the ability to circumvent causing immunogenicity and hepatotoxicity [173]. Wan et al. [62] proposed a novel method for treating liver diseases by utilizing the RNA‐carrying capacity of engineered EVs. They developed EV ribonucleic acids and, upon systemic injection, observed a significant reduction in central lobular cell necrosis and hyperemia in a model of acute liver injury, as well as the inhibition of acetaminophen‐induced acute liver injury and reduced mortality. Encouragingly, in a model of liver fibrosis, this method was also found to decrease the progression of chronic liver fibrosis. An additional study demonstrated that EVs modified with cationic amylopectin can more precisely target the liver and exhibit improved efficacy in treating liver injury [174]. These findings indicate that engineered EVs hold substantial therapeutic potential for the treatment of liver diseases.
The potential of peptide‐modified EVs to treat cardiac tissues has been examined. EVs carrying cardiac tissue‐specific peptides have shown improved cardiac targeting [175]. In another study, the ischemic myocardial targeting peptide was incorporated into the EV membrane protein LAMP‐2B derived from MSCs through genetic engineering [176]. After being intravenously injected into mouse models, these modified EVs were shown to aggregate higher in the ischemic heart region than unmodified EVs. Additionally, in a mouse model of myocardial infarction, these modified EVs significantly reduced inflammation and cardiomyocyte apoptosis, promoted angiogenesis, and enhanced cardiac function (Figure 6) [176].
FIGURE 6.
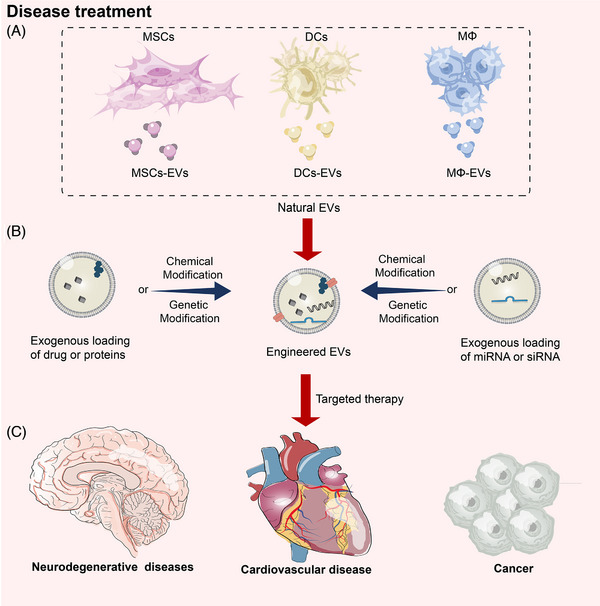
Engineered EVs from different cell sources can be used to treat different diseases. (A) Mesenchymal stem cells, dendritic cells, and macrophages are a few examples of cells that may be employed to make EVs. (B) Each kind of cell has unique qualities that make it appropriate for the treatment of various disorders. EVs from various sources may be created to load therapeutic modalities, including medications, nucleic acids, and proteins, successfully into modified EVs. (C) The recipient cells may be chosen and given the modified EVs. The payload within the modified EVs is finally released for the precise therapy of numerous illnesses, including cancer, neurological disorders, and cardiovascular diseases, once it has been ingested and internalized by the recipient cells. DCs, dendritic cells; MΦ, macrophage cells; MSCs mesenchymal stem cells; MSCs‐EVs, external vesicles derived from mesenchymal stem cells miRNA, microRNA; siRNA, small/short interfering RNA
7. APPLICATION OF ENGINEERED EVS IN OVERCOMING DRUG RESISTANCE
The emergence of drug resistance has become a major problem that needs to be solved in current tumor treatment. Tumor drug resistance is divided into congenital acquisition and acquired acquisition. Among them, acquired acquisition is more common, and the most common drug resistance mechanism includes pro‐survival signals, genetic or epigenetic upregulation of conduction and inhibition of apoptotic pathways, drug inactivation or alteration of drug target molecules, overexpression of multidrug resistance proteins and increased efflux pump transport or drug export [177]. In recent years, many studies have found that tumor‐derived EVs can mediate communication not only between tumor cells but also between cancer cells and stromal cells in the TME, leading to the spread of drug resistance and tumor progression [178, 179, 180]. EVs, because of their low immunogenicity and biocompatibility, can act as an “invisible cloak” for incorporating therapeutic agents and are not cleared by the monocyte‐macrophage system [181]. They also have good tissue penetration capabilities (such as the ability to cross the BBB) and endocytic pathways, acting as a “Trojan horse” to easily deliver therapeutic agents to specific cancer cells [182, 183]. Therefore, more and more studies focus on EVs as the preferred transport system for reversing tumor drug resistance. Some studies have used the EVs secreted by macrophages to load paclitaxel [169, 184]. These EVs can hide paclitaxel in circulation and bind to tumor cells effectively under acidic conditions, so that paclitaxel can induce internalization into cancer cells. Compared with paclitaxel alone, EVs loaded with paclitaxel significantly increased cytotoxicity in drug‐resistant cell lines. Subsequently, a strong anti‐tumor response was observed in the lung metastatic cancer model [148]. In this study, the mechanisms by which EVs are able to reverse drug resistance may be (1) preferential accumulation of EVs in cancer cells, (2) efficient delivery of incorporated paclitaxel into target cancer cells by EVs, and (3) bypasses P‐glycoprotein (P‐gp)‐mediated drug efflux through endocytic mechanisms. We prefer that EVs bypass the P‐gp efflux mechanism and cause it to accumulate in drug‐resistant cells, because regardless of the systemic or oral administration route, the purpose of treatment is to deliver the drug inside the tumor. Whether the drug is successfully taken up by the cells and achieving effective concentrations is a determining factor in drug efficacy and treatment success. Under the condition of low pH in the TME, EVs are more easily taken up by cancer cells, directly bypassing P‐gp through the endocytic pathway of cells and releasing therapeutic drugs in the cell body, thereby achieving the effect of treating tumors [185, 186]. Increasing evidence shows that ncRNAs, such as miRNA and siRNA, are also closely related to tumor drug resistance [187, 188]. Among them, miR‐21 is related to resistance to multiple chemotherapies in colorectal cancer [189, 190]. Therefore, Liang et al. [191] developed EVs which can introduce miR‐21 inhibitors and chemotherapeutic drugs into cells and mouse models resistant to 5‐fluorouracil (5‐FU). Compared with a single dose of miR‐21 inhibitors or 5‐FU, the engineered EVs not only successfully reversed the drug resistance to 5‐FU but also significantly increased its cytotoxicity. Similarly, siRNA has been shown to downregulate key immune transcription factors in tumors, leading to the reversal of drug resistance and induction of anti‐neoplastic effects [192]. To introduce glucose‐regulated protein 78 (GRP78) siRNA into hepatoma cells, effectively reduce the expression of GRP78, and increase the sensitivity of cancer cells resistant to sorafenib, Li et al. [193] used EVs released by bone marrow MSCs. EVs carrying anti‐miR‐214, a miRNA associated with MDR, have also been shown to effectively reverse resistance to cisplatin in gastric cancer and inhibit tumor growth. The formation of drug resistance in tumors is diverse and complex, but just like the “barrel effect”, targeting its key factors can also reverse tumor drug resistance. Similarly, Zhang et al. [194] found that microsomal triglyceride transfer protein (MTTP) can promote resistance of colorectal cancer cells to oxaliplatin by inhibiting iron death through the MTTP/proline‐rich acidic protein 1/zinc finger E‐box binding homeobox 1 axis. When MTTP‐knockdown EVs were used, it was found that drug resistance could be reversed.
The TME is also critical to the formation of tumor drug resistance. The TME can inhibit the death of tumor cells through an immune response, yet they can secrete related factors to promote tumor growth, invasion, and even drug resistance [195]. Although there are few studies on engineering EVs to suppress the microenvironment and overcome drug resistance, there is still a great promise for researches in this field. After all, the tumor and its microenvironment complement each other.
8. CONCLUSIONS AND PROSPECTS
The advent of precision therapy has ushered in a new era in drug delivery. Because of their favorable biological distribution, biocompatibility, and minimal immunogenicity, EVs provide an ideal possibility for EV‐based medication delivery. Engineered EVs also act as a means of transport for small‐molecule drugs and nucleic acids by prolonging their half‐life, decreasing clearance levels, and safeguarding payloads from degradation or inactivation. Furthermore, engineered EVs can be surface‐modified to promote their specific accumulation at the target site, reduce off‐target effects, and surmount drug resistance mechanisms such as P‐gp, thereby facilitating effective treatment of tumors and other diseases. Additionally, the crucial advantage of EVs is their ability to ferry drugs across challenging membranes (such as the BBB), which is not feasible with nanotechnology. However, numerous hurdles exist in the clinical application of EVs. Primarily, the impact of surface modification on EV stability, cellular uptake pathways, and in vivo tissue distribution needs to be elucidated. Moreover, the loading efficiency of therapeutically effective drugs is still inadequate, and therefore, improving it is a primary focus of future research. Finally, although multiple methods for EV extraction exist, most are arduous and costly; thus, realizing large‐scale production of EVs for clinical use is still a significant challenge.
In conclusion, despite some difficulties, modified EVs provide novel therapeutic opportunities for the treatment of cancer and other disorders, as well as a potential therapeutic strategy to overcome tumor drug resistance. We believe that these challenges can be effectively resolved and that engineered EVs will shine as a drug delivery system in clinical treatment.
AUTHOR CONTRIBUTIONS
Chen Ming‐Kun: Writing original draft. Chen Zi‐Xian: Writing original draft. Cai Mao‐Ping: Writing, reviewing, editing and visualization. Chen Hong: Reviewing and reediting. Zhao Shan‐Chao and Chen Zhuang‐Fei: Reviewing, supervision and funding acquisition. All authors contributed to the writing and revision of the manuscript, and approved its submission.
COMPETING INTEREST
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable
CONSENT FOR PUBLICATION
Not applicable
AVAILABILITY OF DATA AND MATERIAL
Not applicable
ACKNOWLEDGEMENTS
This work was supported by grants from the National Natural Science Foundation of China (No. 81972394 and 82272856), the Guang Dong Basic and Applied Basic Research Foundation (No. 2022A1515010437), and the President Foundation of The Third Affiliated Hospital of Southern Medical University (No. YM2021010, No. YM2021011), and the Scientific and Technological Support Project of Guizhou Province (Qian Ke He Zhi Cheng 2023] Yi Ban 262)
Ming‐Kun C, Zi‐Xian C, Mao‐Ping C, Hong C, Zhuang‐Fei C, Shan‐Chao Z. Engineered extracellular vesicles: A new approach for targeted therapy of tumors and overcoming drug resistance. Cancer Commun. 2024;44:205–225. 10.1002/cac2.12518
Chen Ming‐Kun, Chen Zi‐Xian and Cai Mao‐Ping contributed equally to this work.
Contributor Information
Chen Zhuang‐Fei, Email: chenzhuangfei@126.com.
Zhao Shan‐Chao, Email: lulululu@smu.edu.cn.
REFERENCES
- 1. Miller KD, Nogueira L, Devasia T, Mariotto AB, Yabroff KR, Jemal A, et al. Cancer treatment and survivorship statistics, 2022. CA Cancer J Clin. 2022;72(5):409–436. [DOI] [PubMed] [Google Scholar]
- 2. Chen E, Abu‐Sbeih H, Thirumurthi S, Mallepally N, Khurana S, Wei D, et al. Clinical characteristics of colitis induced by taxane‐based chemotherapy. Ann Gastroenterol. 2020;33(1):59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Palukuri NR, Yedla RP, Bala SC, Kuruva SP, Chennamaneni R, Konatam ML, et al. Incidence of febrile neutropenia with commonly used chemotherapy regimen in localized breast cancer. South Asian J Cancer. 2020;9(1):4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu D, Yang F, Xiong F, Gu N. The smart drug delivery system and its clinical potential. Theranostics. 2016;6(9):1306–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsumura Y. The drug discovery by nanomedicine and its clinical experience. Jpn J Clin Oncol. 2014;44(6):515–525. [DOI] [PubMed] [Google Scholar]
- 6. Haque S, Whittaker MR, McIntosh MP, Pouton CW, Kaminskas LM. Disposition and safety of inhaled biodegradable nanomedicines: Opportunities and challenges. Nanomedicine. 2016;12(6):1703–1724. [DOI] [PubMed] [Google Scholar]
- 7. Vader P, Mol EA, Pasterkamp G, Schiffelers RM. Extracellular vesicles for drug delivery. Adv Drug Deliv Rev. 2016;106(Pt A):148–156. [DOI] [PubMed] [Google Scholar]
- 8. Batrakova EV, Kim MS. Using exosomes, naturally‐equipped nanocarriers, for drug delivery. J Control Release. 2015;219:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bang C, Thum T. Exosomes: New players in cell‐cell communication. Int J Biochem Cell Biol. 2012;44(11):2060–2064. [DOI] [PubMed] [Google Scholar]
- 10. Morse MA, Garst J, Osada T, Khan S, Hobeika A, Clay TM, et al. A phase I study of dexosome immunotherapy in patients with advanced non‐small cell lung cancer. J Transl Med. 2005;3(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dai S, Wei D, Wu Z, Zhou X, Wei X, Huang H, et al. Phase I clinical trial of autologous ascites‐derived exosomes combined with GM‐CSF for colorectal cancer. Mol Ther. 2008;16(4):782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Morishita M, Takahashi Y, Matsumoto A, Nishikawa M, Takakura Y. Exosome‐based tumor antigens‐adjuvant co‐delivery utilizing genetically engineered tumor cell‐derived exosomes with immunostimulatory CpG DNA. Biomaterials. 2016;111:55–65. [DOI] [PubMed] [Google Scholar]
- 13. Urbanelli L, Buratta S, Sagini K, Ferrara G, Lanni M, Emiliani C. Exosome‐based strategies for Diagnosis and Therapy. Recent Pat CNS Drug Discov. 2015;10(1):10–27. [DOI] [PubMed] [Google Scholar]
- 14. Kou L, Bhutia YD, Yao Q, He Z, Sun J, Ganapathy V. Transporter‐Guided delivery of nanoparticles to improve drug permeation across cellular barriers and drug exposure to selective cell types. Front Pharmacol. 2018;9:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blanco E, Shen H, Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat Biotechnol. 2015;33(9):941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ngoune R, Peters A, von Elverfeldt D, Winkler K, Pütz G. Accumulating nanoparticles by EPR: A route of no return. J Control Release. 2016;238:58–70. [DOI] [PubMed] [Google Scholar]
- 17. Anselmo AC, Mitragotri S. Nanoparticles in the clinic. Bioeng Transl Med. 2016;1(1):10–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen G, Wang Y, Xie R, Gong S. Tumor‐targeted pH/redox dual‐sensitive unimolecular nanoparticles for efficient siRNA delivery. J Control Release. 2017;259:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mohammad F, Al‐Lohedan HA. Luteinizing hormone‐releasing hormone targeted superparamagnetic gold nanoshells for a combination therapy of hyperthermia and controlled drug delivery. Mater Sci Eng C Mater Biol Appl. 2017;76:692–700. [DOI] [PubMed] [Google Scholar]
- 20. Qiao Y, Wan J, Zhou L, Ma W, Yang Y, Luo W, et al. Stimuli‐responsive nanotherapeutics for precision drug delivery and cancer therapy. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019;11(1):e1527. [DOI] [PubMed] [Google Scholar]
- 21. Lv T, Yu T, Fang Y, Zhang S, Jiang M, Zhang H, et al. Role of generation on folic acid‐modified poly(amidoamine) dendrimers for targeted delivery of baicalin to cancer cells. Mater Sci Eng C Mater Biol Appl. 2017;75:182–190. [DOI] [PubMed] [Google Scholar]
- 22. Xu L, Yeudall WA, Yang H. Folic acid‐decorated polyamidoamine dendrimer exhibits high tumor uptake and sustained highly localized retention in solid tumors: Its utility for local siRNA delivery. Acta Biomater. 2017;57:251–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muralidharan R, Babu A, Amreddy N, Basalingappa K, Mehta M, Chen A, et al. Folate receptor‐targeted nanoparticle delivery of HuR‐RNAi suppresses lung cancer cell proliferation and migration. J Nanobiotechnology. 2016;14(1):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daniels TR, Bernabeu E, Rodríguez JA, Patel S, Kozman M, Chiappetta DA, et al. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim Biophys Acta. 2012;1820(3):291–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu L, Wei Y, Zhai S, Chen Q, Xing D. Dihydroartemisinin and transferrin dual‐dressed nano‐graphene oxide for a pH‐triggered chemotherapy. Biomaterials. 2015;62:35–46. [DOI] [PubMed] [Google Scholar]
- 26. Pant K, Neuber C, Zarschler K, Wodtke J, Meister S, Haag R, et al. Active targeting of dendritic polyglycerols for diagnostic cancer imaging. Small. 2020;16(7):e1905013. [DOI] [PubMed] [Google Scholar]
- 27. Nasrollahi F, Koh YR, Chen P, Varshosaz J, Khodadadi AA, Lim S. Targeting graphene quantum dots to epidermal growth factor receptor for delivery of cisplatin and cellular imaging. Mater Sci Eng C Mater Biol Appl. 2019;94:247–257. [DOI] [PubMed] [Google Scholar]
- 28. Agnello L, Tortorella S, d'Argenio A, Carbone C, Camorani S, Locatelli E, et al. Optimizing cisplatin delivery to triple‐negative breast cancer through novel EGFR aptamer‐conjugated polymeric nanovectors. J Exp Clin Cancer Res. 2021;40(1):239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu Z, Xiang J. Aptamers, the nucleic acid antibodies, in cancer therapy. Int J Mol Sci. 2020;21(8):2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, et al. Targeted nanoparticle‐aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci U S A. 2006;103(16):6315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu X, Ding B, Gao J, Wang H, Fan W, Wang X, et al. Second‐generation aptamer‐conjugated PSMA‐targeted delivery system for prostate cancer therapy. Int J Nanomedicine. 2011;6:1747–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu L, Liu F, Yin L, Wang F, Shi H, Zhao Q, et al. The establishment of polypeptide PSMA‐targeted chimeric antigen receptor‐engineered natural killer cells for castration‐resistant prostate cancer and the induction of ferroptosis‐related cell death. Cancer Commun (Lond). 2022;42(8):768–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tassev DV, Cheng M, Cheung NK. Retargeting NK92 cells using an HLA‐A2‐restricted, EBNA3C‐specific chimeric antigen receptor. Cancer Gene Ther. 2012;19(2):84–100. [DOI] [PubMed] [Google Scholar]
- 34. Mitragotri S, Lammers T, Bae YH, Schwendeman S, De Smedt S, Leroux JC, et al. Drug delivery research for the future: Expanding the nano horizons and beyond. J Control Release. 2017;246:183–184. [DOI] [PubMed] [Google Scholar]
- 35. Zolnik BS, González‐Fernández A, Sadrieh N, Dobrovolskaia MA. Nanoparticles and the immune system. Endocrinology. 2010;151(2):458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu Y, Hardie J, Zhang X, Rotello VM. Effects of engineered nanoparticles on the innate immune system. Semin Immunol. 2017;34:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szebeni J, Storm G. Complement activation as a bioequivalence issue relevant to the development of generic liposomes and other nanoparticulate drugs. Biochem Biophys Res Commun. 2015;468(3):490–497. [DOI] [PubMed] [Google Scholar]
- 38. Szebeni J, Moghimi SM. Liposome triggering of innate immune responses: A perspective on benefits and adverse reactions. J Liposome Res. 2009;19(2):85–90. [DOI] [PubMed] [Google Scholar]
- 39. Laverman P, Carstens MG, Boerman OC, Dams ET, Oyen WJ, van Rooijen N, et al. Factors affecting the accelerated blood clearance of polyethylene glycol‐liposomes upon repeated injection. J Pharmacol Exp Ther. 2001;298(2):607–612. [PubMed] [Google Scholar]
- 40. Schellekens H, Hennink WE, Brinks V. The immunogenicity of polyethylene glycol: Facts and fiction. Pharmaceutical Research. 2013;30(7):1729–1734. [DOI] [PubMed] [Google Scholar]
- 41. Gabizon AA. Stealth liposomes and tumor targeting: One step further in the quest for the magic bullet. Clin Cancer Res. 2001;7(2):223–225. [PubMed] [Google Scholar]
- 42. Tinkle S, McNeil SE, Mühlebach S, Bawa R, Borchard G, Barenholz YC, et al. Nanomedicines: Addressing the scientific and regulatory gap. Ann N Y Acad Sci. 2014;1313:35–56. [DOI] [PubMed] [Google Scholar]
- 43. Grainger DW. Connecting drug delivery reality to smart materials design. Int J Pharm. 2013;454(1):521–524. [DOI] [PubMed] [Google Scholar]
- 44. Nyström AM, Fadeel B. Safety assessment of nanomaterials: Implications for nanomedicine. J Control Release. 2012;161(2):403–408. [DOI] [PubMed] [Google Scholar]
- 45. van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64(3):676–705. [DOI] [PubMed] [Google Scholar]
- 46. Evers MJW, van de Wakker SI, de Groot EM, de Jong OG, Gitz‐François JJJ, Seinen CS, et al. Functional siRNA delivery by extracellular vesicle‐liposome hybrid nanoparticles. Adv Healthc Mater. 2022;11(5):e2101202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roerig J, Mitrach F, Schmid M, Hause G, Hacker MC, Wölk C, et al. Synergistic siRNA loading of extracellular vesicles enables functional delivery into cells. Small Methods. 2022;6(12):e2201001. [DOI] [PubMed] [Google Scholar]
- 48. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell. 1983;33(3):967–978. [DOI] [PubMed] [Google Scholar]
- 49. Jia S, Zocco D, Samuels ML, Chou MF, Chammas R, Skog J, et al. Emerging technologies in extracellular vesicle‐based molecular diagnostics. Expert Rev Mol Diagn. 2014;14(3):307–321. [DOI] [PubMed] [Google Scholar]
- 50. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: Current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820(7):940–948. [DOI] [PubMed] [Google Scholar]
- 51. Sahoo S, Klychko E, Thorne T, Misener S, Schultz KM, Millay M, et al. Exosomes from human CD34(+) stem cells mediate their proangiogenic paracrine activity. Circ Res. 2011;109(7):724–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, et al. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J Transl Med. 2011;9:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101(36):13368–13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. van Dommelen SM, Vader P, Lakhal S, Kooijmans SA, van Solinge WW, Wood MJ, et al. Microvesicles and exosomes: Opportunities for cell‐derived membrane vesicles in drug delivery. J Control Release. 2012;161(2):635–644. [DOI] [PubMed] [Google Scholar]
- 56. Smyth T, Kullberg M, Malik N, Smith‐Jones P, Graner MW, Anchordoquy TJ. Biodistribution and delivery efficiency of unmodified tumor‐derived exosomes. J Control Release. 2015;199:145–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. He C, Zheng S, Luo Y, Wang B. Exosome theranostics: Biology and translational medicine. Theranostics. 2018;8(1):237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rufino‐Ramos D, Albuquerque PR, Carmona V, Perfeito R, Nobre RJ, Pereira de Almeida L. Extracellular vesicles: Novel promising delivery systems for therapy of brain diseases. J Control Release. 2017;262:247–258. [DOI] [PubMed] [Google Scholar]
- 59. Tian T, Zhang HX, He CP, Fan S, Zhu YL, Qi C, et al. Surface functionalized exosomes as targeted drug delivery vehicles for cerebral ischemia therapy. Biomaterials. 2018;150:137–149. [DOI] [PubMed] [Google Scholar]
- 60. Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, et al. Protein typing of circulating microvesicles allows real‐time monitoring of glioblastoma therapy. Nat Med. 2012;18(12):1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wan T, Zhong J, Pan Q, Zhou T, Ping Y, Liu X. Exosome‐mediated delivery of Cas9 ribonucleoprotein complexes for tissue‐specific gene therapy of liver diseases. Sci Adv. 2022;8(37):eabp9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, et al. Reassessment of exosome composition. Cell. 2019;177(2):428–445. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wu Y, Deng W, Klinke DJ 2nd. Exosomes: Improved methods to characterize their morphology, RNA content, and surface protein biomarkers. Analyst. 2015;140(19):6631–6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J Extracell Vesicles. 2015;4:25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Islam MK, Syed P, Lehtinen L, Leivo J, Gidwani K, Wittfooth S, et al. A nanoparticle‐based approach for the detection of extracellular vesicles. Sci Rep. 2019;9(1):10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Livshits MA, Khomyakova E, Evtushenko EG, Lazarev VN, Kulemin NA, Semina SE, et al. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci Rep. 2015;5:17319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Colao IL, Corteling R, Bracewell D, Wall I. Manufacturing exosomes: A promising therapeutic platform. Trends Mol Med. 2018;24(3):242–256. [DOI] [PubMed] [Google Scholar]
- 69. Yang XX, Sun C, Wang L, Guo XL. New insight into isolation, identification techniques and medical applications of exosomes. J Control Release. 2019;308:119–129. [DOI] [PubMed] [Google Scholar]
- 70. Vergauwen G, Dhondt B, Van Deun J, De Smedt E, Berx G, Timmerman E, et al. Confounding factors of ultrafiltration and protein analysis in extracellular vesicle research. Sci Rep. 2017;7(1):2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gurunathan S, Kang MH, Jeyaraj M, Qasim M, Kim JH. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells. 2019;8(4):307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang Y, Bi J, Huang J, Tang Y, Du S, Li P. Exosome: A review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int J Nanomedicine. 2020;15:6917–6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang D, Zhang W, Zhang H, Zhang F, Chen L, Ma L, et al. Progress, opportunity, and perspective on exosome isolation—efforts for efficient exosome‐based theranostics. Theranostics. 2020;10(8):3684–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fitzgerald J, Leonard P, Darcy E, Sharma S, O'Kennedy R. Immunoaffinity chromatography: Concepts and applications. Methods Mol Biol. 2017;1485:27–51. [DOI] [PubMed] [Google Scholar]
- 75. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7(3):789–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Benedikter BJ, Bouwman FG, Vajen T, Heinzmann ACA, Grauls G, Mariman EC, et al. Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Sci Rep. 2017;7(1):15297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shu S, Allen CL, Benjamin‐Davalos S, Koroleva M, MacFarland D, Minderman H, et al. A rapid exosome isolation using ultrafiltration and size exclusion chromatography (REIUS) method for exosome isolation from melanoma cell lines. Methods Mol Biol. 2021;2265:289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Guan S, Yu H, Yan G, Gao M, Sun W, Zhang X. Characterization of urinary exosomes purified with size exclusion chromatography and ultracentrifugation. J Proteome Res. 2020;19(6):2217–2225. [DOI] [PubMed] [Google Scholar]
- 79. Mittelbrunn M, Sánchez‐Madrid F. Intercellular communication: Diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol. 2012;13(5):328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Prada I, Meldolesi J. Binding and fusion of extracellular vesicles to the plasma membrane of their cell targets. Int J Mol Sci. 2016;17(8):1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tian T, Wang Y, Wang H, Zhu Z, Xiao Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live‐cell microscopy. J Cell Biochem. 2010;111(2):488–496. [DOI] [PubMed] [Google Scholar]
- 82. Liu Q, Rojas‐Canales DM, Divito SJ, Shufesky WJ, Stolz DB, Erdos G, et al. Donor dendritic cell‐derived exosomes promote allograft‐targeting immune response. J Clin Invest. 2016;126(8):2805–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Coleman BM, Hill AF. Extracellular vesicles–Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Semin Cell Dev Biol. 2015;40:89–96. [DOI] [PubMed] [Google Scholar]
- 84. Rajendran L, Bali J, Barr MM, Court FA, Krämer‐Albers EM, Picou F, et al. Emerging roles of extracellular vesicles in the nervous system. J Neurosci. 2014;34(46):15482–15489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Limoni SK, Moghadam MF, Moazzeni SM, Gomari H, Salimi F. Engineered exosomes for targeted transfer of siRNA to HER2 positive breast cancer cells. Appl Biochem Biotechnol. 2019;187(1):352–364. [DOI] [PubMed] [Google Scholar]
- 86. Barile L, Vassalli G. Exosomes: Therapy delivery tools and biomarkers of diseases. Pharmacol Ther. 2017;174:63–78. [DOI] [PubMed] [Google Scholar]
- 87. Zou J, Shi M, Liu X, Jin C, Xing X, Qiu L, et al. Aptamer‐Functionalized exosomes: Elucidating the cellular uptake mechanism and the potential for cancer‐targeted chemotherapy. Anal Chem. 2019;91(3):2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Smyth T, Petrova K, Payton NM, Persaud I, Redzic JS, Graner MW, et al. Surface functionalization of exosomes using click chemistry. Bioconjug Chem. 2014;25(10):1777–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jia G, Han Y, An Y, Ding Y, He C, Wang X, et al. NRP‐1 targeted and cargo‐loaded exosomes facilitate simultaneous imaging and therapy of glioma in vitro and in vivo. Biomaterials. 2018;178:302–316. [DOI] [PubMed] [Google Scholar]
- 90. Abumiya T, Lucero J, Heo JH, Tagaya M, Koziol JA, Copeland BR, et al. Activated microvessels express vascular endothelial growth factor and integrin alpha(v)beta3 during focal cerebral ischemia. J Cereb Blood Flow Metab. 1999;19(9):1038–1050. [DOI] [PubMed] [Google Scholar]
- 91. Haubner R, Wester HJ, Burkhart F, Senekowitsch‐Schmidtke R, Weber W, Goodman SL, et al. Glycosylated RGD‐containing peptides: Tracer for tumor targeting and angiogenesis imaging with improved biokinetics. J Nucl Med. 2001;42(2):326–336. [PubMed] [Google Scholar]
- 92. Elward K, Gasque P. “Eat me” and “don't eat me” signals govern the innate immune response and tissue repair in the CNS: Emphasis on the critical role of the complement system. Mol Immunol. 2003;40(2‐4):85–94. [DOI] [PubMed] [Google Scholar]
- 93. Chao MP, Weissman IL, Majeti R. The CD47‐SIRPα pathway in cancer immune evasion and potential therapeutic implications. Curr Opin Immunol. 2012;24(2):225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nie W, Wu G, Zhang J, Huang LL, Ding J, Jiang A, et al. Responsive exosome nano‐bioconjugates for synergistic cancer therapy. Angew Chem Int Ed Engl. 2020;59(5):2018–2022. [DOI] [PubMed] [Google Scholar]
- 95. Ji C, Wei J, Zhang L, Hou X, Tan J, Yuan Q, et al. Aptamer‐Protein interactions: From regulation to biomolecular detection. Chem Rev. 2023;123(22):12471–12506. [DOI] [PubMed] [Google Scholar]
- 96. Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103(32):11838–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Fang X, Tan W. Aptamers generated from cell‐SELEX for molecular medicine: A chemical biology approach. Acc Chem Res. 2010;43(1):48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang Y, Chen X, Tian B, Liu J, Yang L, Zeng L, et al. Nucleolin‐targeted extracellular vesicles as a versatile platform for biologics delivery to breast cancer. Theranostics. 2017;7(5):1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pi F, Binzel DW, Lee TJ, Li Z, Sun M, Rychahou P, et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat Nanotechnol. 2018;13(1):82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Qi H, Liu C, Long L, Ren Y, Zhang S, Chang X, et al. Blood exosomes endowed with magnetic and targeting properties for cancer therapy. ACS Nano. 2016;10(3):3323–3333. [DOI] [PubMed] [Google Scholar]
- 101. Nakase I, Futaki S. Combined treatment with a pH‐sensitive fusogenic peptide and cationic lipids achieves enhanced cytosolic delivery of exosomes. Sci Rep. 2015;5:10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kooijmans SA, Aleza CG, Roffler SR, van Solinge WW, Vader P, Schiffelers RM. Display of GPI‐anchored anti‐EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J Extracell Vesicles. 2016;5:31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Akbari A, Nazari‐Khanamiri F, Ahmadi M, Shoaran M, Rezaie J. Engineered exosomes for tumor‐targeted drug delivery: A focus on genetic and chemical functionalization. Pharmaceutics. 2022;15(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Luan X, Sansanaphongpricha K, Myers I, Chen H, Yuan H, Sun D. Engineering exosomes as refined biological nanoplatforms for drug delivery. Acta Pharmacol Sin. 2017;38(6):754–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Salunkhe S, Dheeraj BM, Chitkara D, Mittal A. Surface functionalization of exosomes for target‐specific delivery and in vivo imaging & tracking: Strategies and significance. J Control Release. 2020;326:599–614. [DOI] [PubMed] [Google Scholar]
- 106. Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. [DOI] [PubMed] [Google Scholar]
- 107. Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448(7149):39–43. [DOI] [PubMed] [Google Scholar]
- 108. Alvarez‐Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–345. [DOI] [PubMed] [Google Scholar]
- 109. Wilson HL, Ni K, O'Neill HC. Identification of progenitor cells in long‐term spleen stromal cultures that produce immature dendritic cells. Proc Natl Acad Sci U S A. 2000;97(9):4784–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Quah BJ, O'Neill HC. The immunogenicity of dendritic cell‐derived exosomes. Blood Cells Mol Dis. 2005;35(2):94–110. [DOI] [PubMed] [Google Scholar]
- 111. Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–2390. [DOI] [PubMed] [Google Scholar]
- 112. Bai J, Duan J, Liu R, Du Y, Luo Q, Cui Y, et al. Engineered targeting tLyp‐1 exosomes as gene therapy vectors for efficient delivery of siRNA into lung cancer cells. Asian J Pharm Sci. 2020;15(4):461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Teesalu T, Sugahara KN, Kotamraju VR, Ruoslahti E. C‐end rule peptides mediate neuropilin‐1‐dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci U S A. 2009;106(38):16157–16162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kawakami T, Tokunaga T, Hatanaka H, Kijima H, Yamazaki H, Abe Y, et al. Neuropilin 1 and neuropilin 2 co‐expression is significantly correlated with increased vascularity and poor prognosis in nonsmall cell lung carcinoma. Cancer. 2002;95(10):2196–2201. [DOI] [PubMed] [Google Scholar]
- 115. Hung ME, Leonard JN. Stabilization of exosome‐targeting peptides via engineered glycosylation. J Biol Chem. 2015;290(13):8166–8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Cheng Q, Shi X, Han M, Smbatyan G, Lenz HJ, Zhang Y. Reprogramming exosomes as nanoscale controllers of cellular immunity. J Am Chem Soc. 2018;140(48):16413–16417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Liang G, Kan S, Zhu Y, Feng S, Feng W, Gao S. Engineered exosome‐mediated delivery of functionally active miR‐26a and its enhanced suppression effect in HepG2 cells. Int J Nanomedicine. 2018;13:585–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Ran N, Gao X, Dong X, Li J, Lin C, Geng M, et al. Effects of exosome‐mediated delivery of myostatin propeptide on functional recovery of mdx mice. Biomaterials. 2020;236:119826. [DOI] [PubMed] [Google Scholar]
- 119. Srinivasan S, Vannberg FO, Dixon JB. Lymphatic transport of exosomes as a rapid route of information dissemination to the lymph node. Sci Rep. 2016;6:24436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Armstrong JP, Holme MN, Stevens MM. Re‐Engineering extracellular vesicles as smart nanoscale therapeutics. ACS Nano. 2017;11(1):69–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Pascucci L, Coccè V, Bonomi A, Ami D, Ceccarelli P, Ciusani E, et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J Control Release. 2014;192:262–270. [DOI] [PubMed] [Google Scholar]
- 122. O'Loughlin AJ, Mäger I, de Jong OG, Varela MA, Schiffelers RM, El Andaloussi S, et al. Functional delivery of lipid‐conjugated siRNA by extracellular vesicles. Mol Ther. 2017;25(7):1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Cheng L, Zhang K, Wu S, Cui M, Xu T. Focus on mesenchymal stem cell‐derived exosomes: Opportunities and challenges in cell‐free therapy. Stem Cells Int. 2017;2017:6305295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Liu C, Su C. Design strategies and application progress of therapeutic exosomes. Theranostics. 2019;9(4):1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Villata S, Canta M, Cauda V. EVs and bioengineering: From cellular products to engineered nanomachines. Int J Mol Sci. 2020;21(17):6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Fuhrmann G, Serio A, Mazo M, Nair R, Stevens MM. Active loading into extracellular vesicles significantly improves the cellular uptake and photodynamic effect of porphyrins. J Control Release. 2015;205:35–44. [DOI] [PubMed] [Google Scholar]
- 127. Nawaz M, Heydarkhan‐Hagvall S, Tangruksa B, González‐King Garibotti H, Jing Y, Maugeri M, et al. Lipid nanoparticles deliver the therapeutic VEGFA mRNA in vitro and in vivo and transform extracellular vesicles for their functional extensions. Adv Sci (Weinh). 2023;10(12):e2206187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Anguela XM, High KA. Entering the modern era of gene therapy. Annu Rev Med. 2019;70:273–288. [DOI] [PubMed] [Google Scholar]
- 129. Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Winkle M, El‐Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics—challenges and potential solutions. Nat Rev Drug Discov. 2021;20(8):629–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kara G, Calin GA, Ozpolat B. RNAi‐based therapeutics and tumor targeted delivery in cancer. Adv Drug Deliv Rev. 2022;182:114113. [DOI] [PubMed] [Google Scholar]
- 132. Tekedereli I, Alpay SN, Akar U, Yuca E, Ayugo‐Rodriguez C, Han HD, et al. Therapeutic silencing of Bcl‐2 by systemically administered siRNA nanotherapeutics inhibits tumor growth by autophagy and apoptosis and enhances the efficacy of chemotherapy in orthotopic xenograft models of ER (‐) and ER (+) breast cancer. Mol Ther Nucleic Acids. 2013;2(9):e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Reyes‐González JM, Armaiz‐Peña GN, Mangala LS, Valiyeva F, Ivan C, Pradeep S, et al. Targeting c‐MYC in platinum‐resistant ovarian cancer. Mol Cancer Ther. 2015;14(10):2260–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Pecot CV, Wu SY, Bellister S, Filant J, Rupaimoole R, Hisamatsu T, et al. Therapeutic silencing of KRAS using systemically delivered siRNAs. Mol Cancer Ther. 2014;13(12):2876–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Akao Y, Iio A, Itoh T, Noguchi S, Itoh Y, Ohtsuki Y, et al. Microvesicle‐mediated RNA molecule delivery system using monocytes/macrophages. Mol Ther. 2011;19(2):395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Youn K, Park JH, Lee J, Jeong WS, Ho CT, Jun M. The identification of biochanin A as a Potent and selective β‐Site App‐Cleaving Enzyme 1 (Bace1) Inhibitor. Nutrients. 2016;8(10):637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Zhang H, Wang Y, Bai M, Wang J, Zhu K, Liu R, et al. Exosomes serve as nanoparticles to suppress tumor growth and angiogenesis in gastric cancer by delivering hepatocyte growth factor siRNA. Cancer Sci. 2018;109(3):629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Nie H, Xie X, Zhang D, Zhou Y, Li B, Li F, et al. Use of lung‐specific exosomes for miRNA‐126 delivery in non‐small cell lung cancer. Nanoscale. 2020;12(2):877–887. [DOI] [PubMed] [Google Scholar]
- 139. Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Zhu X, Badawi M, Pomeroy S, Sutaria DS, Xie Z, Baek A, et al. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J Extracell Vesicles. 2017;6(1):1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Saleh AF, Lázaro‐Ibáñez E, Forsgard MA, Shatnyeva O, Osteikoetxea X, Karlsson F, et al. Extracellular vesicles induce minimal hepatotoxicity and immunogenicity. Nanoscale. 2019;11(14):6990–7001. [DOI] [PubMed] [Google Scholar]
- 142. Anand P, Thomas SG, Kunnumakkara AB, Sundaram C, Harikumar KB, Sung B, et al. Biological activities of curcumin and its analogues (Congeners) made by man and Mother Nature. Biochem Pharmacol. 2008;76(11):1590–1611. [DOI] [PubMed] [Google Scholar]
- 143. Ravindran J, Prasad S, Aggarwal BB. Curcumin and cancer cells: How many ways can curry kill tumor cells selectively? Aaps j. 2009;11(3):495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: Problems and promises. Mol Pharm. 2007;4(6):807–818. [DOI] [PubMed] [Google Scholar]
- 145. Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C, et al. A novel nanoparticle drug delivery system: The anti‐inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther. 2010;18(9):1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wang M, Yuan Q, Xie L. Mesenchymal stem cell‐based immunomodulation: Properties and clinical application. Stem Cells Int. 2018;2018:3057624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Gomari H, Forouzandeh Moghadam M, Soleimani M, Ghavami M, Khodashenas S. Targeted delivery of doxorubicin to HER2 positive tumor models. Int J Nanomedicine. 2019;14:5679–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, et al. Development of exosome‐encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12(3):655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Yan F, Zhong Z, Wang Y, Feng Y, Mei Z, Li H, et al. Exosome‐based biomimetic nanoparticles targeted to inflamed joints for enhanced treatment of rheumatoid arthritis. J Nanobiotechnology. 2020;18(1):115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Vashisht M, Rani P, Onteru SK, Singh D. Curcumin encapsulated in milk exosomes resists human digestion and possesses enhanced intestinal permeability in vitro. Appl Biochem Biotechnol. 2017;183(3):993–1007. [DOI] [PubMed] [Google Scholar]
- 151. Munagala R, Aqil F, Jeyabalan J, Gupta RC. Bovine milk‐derived exosomes for drug delivery. Cancer Lett. 2016;371(1):48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. [DOI] [PubMed] [Google Scholar]
- 153. Parrales A, Iwakuma T. Targeting oncogenic mutant p53 for cancer therapy. Front Oncol. 2015;5:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Setten RL, Rossi JJ, Han SP. The current state and future directions of RNAi‐based therapeutics. Nat Rev Drug Discov. 2019;18(6):421–446. [DOI] [PubMed] [Google Scholar]
- 155. Roma‐Rodrigues C, Mendes R, Baptista PV, Fernandes AR. Targeting tumor microenvironment for cancer therapy. Int J Mol Sci. 2019;20(4):107753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Hu S, Ma J, Su C, Chen Y, Shu Y, Qi Z, et al. Engineered exosome‐like nanovesicles suppress tumor growth by reprogramming tumor microenvironment and promoting tumor ferroptosis. Acta Biomater. 2021;135:567–581. [DOI] [PubMed] [Google Scholar]
- 157. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour‐associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Chanmee T, Ontong P, Konno K, Itano N. Tumor‐associated macrophages as major players in the tumor microenvironment. Cancers (Basel). 2014;6(3):1670–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Nonnenmacher Y, Hiller K. Biochemistry of proinflammatory macrophage activation. Cell Mol Life Sci. 2018;75(12):2093–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kamerkar S, Leng C, Burenkova O, Jang SC, McCoy C, Zhang K, et al. Exosome‐mediated genetic reprogramming of tumor‐associated macrophages by exoASO‐STAT6 leads to potent monotherapy antitumor activity. Sci Adv. 2022;8(7):eabj7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Gordon S, Martinez FO. Alternative activation of macrophages: Mechanism and functions. Immunity. 2010;32(5):593–604. [DOI] [PubMed] [Google Scholar]
- 162. Deng J, Li J, Sarde A, Lines JL, Lee YC, Qian DC, et al. Hypoxia‐induced VISTA promotes the suppressive function of myeloid‐derived suppressor cells in the tumor microenvironment. Cancer Immunol Res. 2019;7(7):1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Cho SH, Raybuck AL, Stengel K, Wei M, Beck TC, Volanakis E, et al. Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature. 2016;537(7619):234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Brand A, Singer K, Koehl GE, Kolitzus M, Schoenhammer G, Thiel A, et al. LDHA‐associated lactic acid production blunts tumor immunosurveillance by T and NK cells. Cell Metab. 2016;24(5):657–671. [DOI] [PubMed] [Google Scholar]
- 165. Wu T, Liu Y, Cao Y, Liu Z. Engineering macrophage exosome disguised biodegradable nanoplatform for enhanced sonodynamic therapy of glioblastoma. Adv Mater. 2022;34(15):e2110364. [DOI] [PubMed] [Google Scholar]
- 166. Padoan A, Plebani M, Basso D. Inflammation and pancreatic cancer: Focus on metabolism, cytokines, and immunity. Int J Mol Sci. 2019;20(3):676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. De Blander H, Morel AP, Senaratne AP, Ouzounova M, Puisieux A. Cellular plasticity: A route to senescence exit and tumorigenesis. Cancers (Basel). 2021;13(18):4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Jin C, Lagoudas GK, Zhao C, Bullman S, Bhutkar A, Hu B, et al. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176(5):998–1013.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Kim MS, Haney MJ, Zhao Y, Yuan D, Deygen I, Klyachko NL, et al. Engineering macrophage‐derived exosomes for targeted paclitaxel delivery to pulmonary metastases: In vitro and in vivo evaluations. Nanomedicine. 2018;14(1):195–204. [DOI] [PubMed] [Google Scholar]
- 170. Huang L, Rong Y, Tang X, Yi K, Qi P, Hou J, et al. Engineered exosomes as an in situ DC‐primed vaccine to boost antitumor immunity in breast cancer. Mol Cancer. 2022;21(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Lv Q, Cheng L, Lu Y, Zhang X, Wang Y, Deng J, et al. Thermosensitive exosome‐liposome hybrid nanoparticle‐mediated chemoimmunotherapy for improved treatment of metastatic peritoneal cancer. Adv Sci (Weinh). 2020;7(18):2000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Wang J, Tang W, Yang M, Yin Y, Li H, Hu F, et al. Inflammatory tumor microenvironment responsive neutrophil exosomes‐based drug delivery system for targeted glioma therapy. Biomaterials. 2021;273:120784. [DOI] [PubMed] [Google Scholar]
- 173. Ding B, Li T, Zhang J, Zhao L, Zhai G. Advances in liver‐directed gene therapy for hepatocellular carcinoma by non‐viral delivery systems. Curr Gene Ther. 2012;12(2):92–102. [DOI] [PubMed] [Google Scholar]
- 174. Tamura R, Uemoto S, Tabata Y. Augmented liver targeting of exosomes by surface modification with cationized pullulan. Acta Biomater. 2017;57:274–284. [DOI] [PubMed] [Google Scholar]
- 175. Kim H, Yun N, Mun D, Kang JY, Lee SH, Park H, et al. Cardiac‐specific delivery by cardiac tissue‐targeting peptide‐expressing exosomes. Biochem Biophys Res Commun. 2018;499(4):803–808. [DOI] [PubMed] [Google Scholar]
- 176. Wang X, Chen Y, Zhao Z, Meng Q, Yu Y, Sun J, et al. Engineered exosomes with ischemic myocardium‐targeting peptide for targeted therapy in myocardial infarction. J Am Heart Assoc. 2018;7(15):e008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Maacha S, Bhat AA, Jimenez L, Raza A, Haris M, Uddin S, et al. Extracellular vesicles‐mediated intercellular communication: Roles in the tumor microenvironment and anti‐cancer drug resistance. Mol Cancer. 2019;18(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Ifergan I, Scheffer GL, Assaraf YG. Novel extracellular vesicles mediate an ABCG2‐dependent anticancer drug sequestration and resistance. Cancer Res. 2005;65(23):10952–10958. [DOI] [PubMed] [Google Scholar]
- 179. Zhang FF, Zhu YF, Zhao QN, Yang DT, Dong YP, Jiang L, et al. Microvesicles mediate transfer of P‐glycoprotein to paclitaxel‐sensitive A2780 human ovarian cancer cells, conferring paclitaxel‐resistance. Eur J Pharmacol. 2014;738:83–90. [DOI] [PubMed] [Google Scholar]
- 180. Lu JF, Luk F, Gong J, Jaiswal R, Grau GE, Bebawy M. Microparticles mediate MRP1 intercellular transfer and the re‐templating of intrinsic resistance pathways. Pharmacol Res. 2013;76:77–83. [DOI] [PubMed] [Google Scholar]
- 181. Parada N, Romero‐Trujillo A, Georges N, Alcayaga‐Miranda F. Camouflage strategies for therapeutic exosomes evasion from phagocytosis. J Adv Res. 2021;31:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Lin Z, Wu Y, Xu Y, Li G, Li Z, Liu T. Mesenchymal stem cell‐derived exosomes in cancer therapy resistance: Recent advances and therapeutic potential. Mol Cancer. 2022;21(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Avgoulas DI, Tasioulis KS, Papi RM, Pantazaki AA. Therapeutic and diagnostic potential of exosomes as drug delivery systems in brain cancer. Pharmaceutics. 2023;15(5):1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Agrawal AK, Aqil F, Jeyabalan J, Spencer WA, Beck J, Gachuki BW, et al. Milk‐derived exosomes for oral delivery of paclitaxel. Nanomedicine. 2017;13(5):1627–1636. [DOI] [PubMed] [Google Scholar]
- 185. Bach DH, Hong JY, Park HJ, Lee SK. The role of exosomes and miRNAs in drug‐resistance of cancer cells. Int J Cancer. 2017;141(2):220–230. [DOI] [PubMed] [Google Scholar]
- 186. Milman N, Ginini L, Gil Z. Exosomes and their role in tumorigenesis and anticancer drug resistance. Drug Resist Updat. 2019;45:1–12. [DOI] [PubMed] [Google Scholar]
- 187. Pan G, Liu Y, Shang L, Zhou F, Yang S. EMT‐associated microRNAs and their roles in cancer stemness and drug resistance. Cancer Commun (Lond). 2021;41(3):199–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Fatemian T, Othman I, Chowdhury EH. Strategies and validation for siRNA‐based therapeutics for the reversal of multi‐drug resistance in cancer. Drug Discov Today. 2014;19(1):71–78. [DOI] [PubMed] [Google Scholar]
- 189. Mitamura T, Watari H, Wang L, Kanno H, Hassan MK, Miyazaki M, et al. Downregulation of miRNA‐31 induces taxane resistance in ovarian cancer cells through increase of receptor tyrosine kinase MET. Oncogenesis. 2013;2(3):e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF, et al. Involvement of microRNA‐451 in resistance of the MCF‐7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther. 2008;7(7):2152–2159. [DOI] [PubMed] [Google Scholar]
- 191. Liang G, Zhu Y, Ali DJ, Tian T, Xu H, Si K, et al. Engineered exosomes for targeted co‐delivery of miR‐21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology. 2020;18(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Poeck H, Besch R, Maihoefer C, Renn M, Tormo D, Morskaya SS, et al. 5'‐Triphosphate‐siRNA: Turning gene silencing and Rig‐I activation against melanoma. Nat Med. 2008;14(11):1256–1263. [DOI] [PubMed] [Google Scholar]
- 193. Li H, Yang C, Shi Y, Zhao L. Exosomes derived from siRNA against GRP78 modified bone‐marrow‐derived mesenchymal stem cells suppress Sorafenib resistance in hepatocellular carcinoma. J Nanobiotechnology. 2018;16(1):103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Zhang Q, Deng T, Zhang H, Zuo D, Zhu Q, Bai M, et al. Adipocyte‐derived exosomal MTTP suppresses ferroptosis and promotes chemoresistance in colorectal cancer. Adv Sci (Weinh). 2022;9(28):e2203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Erin N, Grahovac J, Brozovic A, Efferth T. Tumor microenvironment and epithelial mesenchymal transition as targets to overcome tumor multidrug resistance. Drug Resist Updat. 2020;53:100715. [DOI] [PubMed] [Google Scholar]
- 196. Zhao S, Xiu G, Wang J, Wen Y, Lu J, Wu B, et al. Engineering exosomes derived from subcutaneous fat MSCs specially promote cartilage repair as miR‐199a‐3p delivery vehicles in Osteoarthritis. J Nanobiotechnology. 2023;21(1):341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Liu J, Ren H, Zhang C, Li J, Qiu Q, Zhang N, et al. Orally‐delivered, cytokine‐engineered extracellular vesicles for targeted treatment of inflammatory bowel disease. Small. 2023;19(50):e2304023. [DOI] [PubMed] [Google Scholar]
- 198. Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Zhang Z, Guo X, Guo X, Yu R, Qian M, Wang S, et al. MicroRNA‐29a‐3p delivery via exosomes derived from engineered human mesenchymal stem cells exerts tumour suppressive effects by inhibiting migration and vasculogenic mimicry in glioma. Aging (Albany NY). 2021;13(4):5055–5068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Liang Q, Bie N, Yong T, Tang K, Shi X, Wei Z, et al. The softness of tumour‐cell‐derived microparticles regulates their drug‐delivery efficiency. Nat Biomed Eng. 2019;3(9):729–740. [DOI] [PubMed] [Google Scholar]
- 201. Lee H, Park H, Noh GJ, Lee ES. pH‐responsive hyaluronate‐anchored extracellular vesicles to promote tumor‐targeted drug delivery. Carbohydr Polym. 2018;202:323–333. [DOI] [PubMed] [Google Scholar]
- 202. McAndrews KM, Xiao F, Chronopoulos A, LeBleu VS, Kugeratski FG, Kalluri R. Exosome‐mediated delivery of CRISPR/Cas9 for targeting of oncogenic Kras(G12D) in pancreatic cancer. Life Sci Alliance. 2021;4(9):e202000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Dar GH, Mendes CC, Kuan WL, Speciale AA, Conceição M, Görgens A, et al. GAPDH controls extracellular vesicle biogenesis and enhances the therapeutic potential of EV mediated siRNA delivery to the brain. Nat Commun. 2021;12(1):6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable


