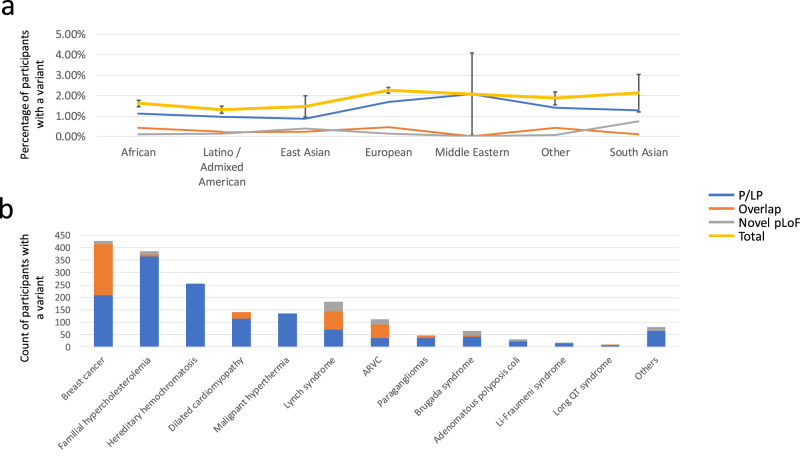
Fig. 1. Pathogenic variants by ancestry.
Using a database of known pathogenic mutations and annotations for rare, pLoF variants, we searched the beta release of the All of Us cohort for pathogenic variants, on the Researcher Workbench. Figure 1a shows the rates of pathogenic variation, broken down by predicted genetic ancestry groups. Error bars show 95% the confidence intervals for the total set of pathogenic variants (including both VIP P/LP variants and rare pLoF). Figure 1b shows the breakdown of pathogenic variants by disease area. The blue line and bar depict the rate of Pathogenic and Likely pathogenic variants, the gray bar the rate of novel, predicted loss of function variants and the orange bar depicts predicted loss of function variants that were known pathogenic variants at the time of analysis. The yellow line shows the total variants in each ancestry group.