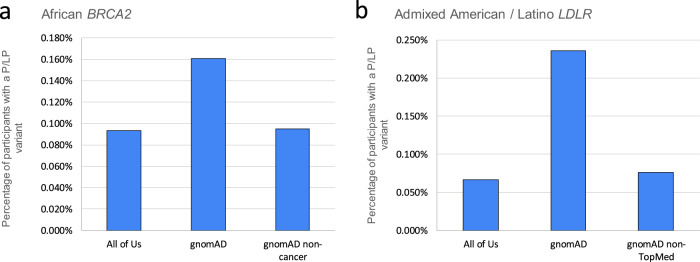
Fig. 3. Comparisons to gnomAD subsets.
Though there is high level concordance between the rates of pathogenic variants in All of Us and gnomAD, in some cases there are differences specific to a gene and ancestry group. For example, in participants with African ancestry, the rate of pathogenic variants in BRCA2 diverges from gnomAD (a). However, when the non-cancer subgroup of gnomAD is used, the rates are much more similar. A similar situation is seen in the Admixed American / Latino ancestry group with LDLR (b). Using the non-TopMed portion of gnomAD brings the rates much closer.