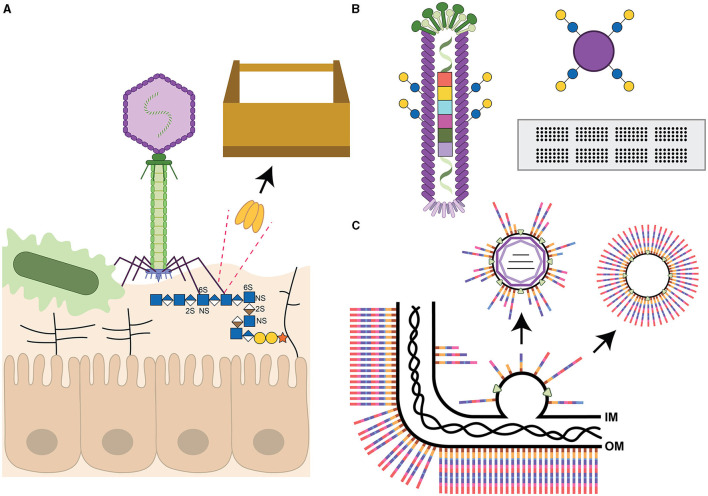
Figure 1.
Summary of bacteriophage components described in this review. (A) T4-like phage (not drawn to scale) with carbohydrate-binding (lectin) tail fibers adhering to the capsular polysaccharides of a microbe, and likely possessing glycoside hydrolase/lyase activity to cleave the receptor to gain access to the bacterial cell wall. The tail fiber is also shown to interact with heparan sulfate glycosaminoglycans (GAG, represented as horizontal lines except for the GAG interacting with the phage) extending from proteins on host epithelial cells (according to Green et al., 2021). The dotted lines highlight one of the receptor binding proteins, depicted as a trimer, as a candidate for the toolbox of reagents useful for glycan detection or cleavage. (B) Filamentous phage covalently modified with glucose and galactose disaccharides as part of the liquid glycan array (LiGA) technology. The phage genome has also been tagged with a barcode to enable identification of adherent phages from deep-sequencing data (according to Sojitra et al., 2021). This glycan display platform is compared to two commonly used methods, Luminex beads modified with the same sugars as shown for the phage, and glycan arrays on glass slides. (C) Assembly of a double-stranded RNA Cystovirus demonstrating that the viral envelope is derived from the host inner membrane (IM) by an unknown self-assembly process and may contain intermediates from bacterial glycoconjugate biosynthesis pathways. The bacterial outer membrane (OM) with a generic full-length polysaccharide is shown with the peptidoglycan layer underneath. The rightmost arrow depicts the possibility of engineering self-assembling lipid vesicles with defined carbohydrate structures. All carbohydrates are drawn using the Symbol Nomenclature for Glycans (Neelamegham et al., 2019). Iduronic acid (divided brown diamond), galactose (yellow circle), glucose (blue circle), glucuronic acid (divided blue diamond), N-acetylglucosamine (blue square), and xylose (orange star). S indicates sites of sulfation with arbitrary linkage site.