Abstract
Thioredoxin-interacting protein (TXNIP) plays a key role in diabetes development and prognosis through its role in pancreatic β-cell dysfunction and death as well as in upregulating the inflammatory response in hyperglycemia. DNA methylation (DNAm) of TXNIP (TXNIP-cg19693031) is associated with the prevalence and incidence of type 2 diabetes (T2D); however, its role in inflammation and its relationship with T2D remain unclear. We aimed to investigate the epigenetic associations of TXNIP-cg19693031 with a panel of inflammatory biomarkers and to examine whether these inflammatory biomarkers modify the association between TXNIP-cg19693031 methylation and diabetes in 218 middle-aged male twins from the Emory Twin Study. We confirmed the association of TXNIP-cg19693031 DNAm with T2D, as well as with HbA1c, insulin and fasting glucose. We found that hypomethylation at TXNIP-cg19693031 is strongly associated with both type 2 diabetes and higher levels of inflammatory biomarkers (VCAM-1, ICAM-1, MMP-2, sRAGE and P-selectin); however, the relationship between TXNIP-cg19693031 and T2D is independent of the levels of these inflammatory biomarkers. Our results suggest that DNA methylation of TXNIP is linked with multiple biological processes, through which the TXNIP may have broad influence on chronic disease risk.
Keywords: DNA methylation, epigenetic epidemiology, inflammation, twin study, TXNIP
Hyperglycemia is considered a primary risk factor for type 2 diabetes (T2D) and its complications (Chen et al., 2008; Mortuza et al., 2013). The fundamental mechanism leading to diabetes is increased metabolic input into cells and induction of oxidative stress. This process occurs via increased formation of reactive oxygen species (ROS; Mortuza et al., 2013), NLRP3 inflammasome activity and apoptosis via mTOR — all pathways in which thioredoxin-interacting protein (TXNIP) plays a major role (Kumar & Mittal, 2018). TXNIP is a ubiquitously expressed protein that inhibits the major cellular antioxidant protein thioredoxin and thus regulates redox-dependent signal pathways, mediates oxidative stress, suppresses cell growth and induces cell apoptosis (Chen et al., 2008). Upregulated TXNIP protein levels were found in pancreatic islets from diabetic mice (Minn et al., 2005) and have been associated with pancreatic beta-cell apoptosis in a human islet microarray study (Minn et al., 2005). TXNIP upregulation has been demonstrated to contribute to the development of both type 1 diabetes and T2D in mice and human isolated islets (Chen et al., 2008; Hui et al., 2008; Khodneva et al., 2016; Yin et al., 2017). Txnip silencing in oxidative tissues (skeletal muscle and hearts) in a mouse model led to improved insulin sensitivity and glucose tolerance (Hui et al., 2008) and attenuated hyperglycemia-induced pancreatic beta-cell apoptosis (Chen et al., 2008). In human studies, downregulation of TXNIP in human islets via administration of calcium channel blockers (Xu et al., 2012), such as the widely used hypertension medication verapamil, was associated with lower levels of fasting blood glucose in diabetic participants (Khodneva et al., 2016; Xu et al., 2012) and significantly reduced incidence of T2D in patients older than 65 years (Yin et al., 2017).
Several studies have reported that overexpression of TXNIP in diabetic rat retinas was linked to inflammation of retinal cells and development of diabetic retinopathy (Perrone et al., 2009; Singh et al., 2017). Increased expression of TXNIP may also contribute to endothelial inflammation and progression of diabetes (Perrone et al., 2009). Recent studies have shown that TXNIP protein-activated gene expression of intercellular adhesion molecule 1 (ICAM-1) (Wongeakin et al., 2014), interleukin 6 (IL-6) and vascular cell adhesion protein 1 (VCAM-1) (Nagai et al., 2007) contribute to inflammation and the development of diabetes.
Epigenome-wide association studies (EWAS) of T2D and hemoglobin A1c (HbA1c) have identified a strong association of DNA methylation (DNAm) site cg1969031 on the TXNIP gene, regardless of ethnic, social and environmental differences in study populations (Chambers et al., 2015; Florath et al., 2016; Kulkarni et al., 2015; Mathur et al., 2019; Meeks et al., 2019; Soriano-Tarraga et al., 2016; Walaszczyk et al., 2018). TXNIP-cg19693031 is located on chromosome 1, base pair position 145441552 (human genome built hg19). Decreased methylation at TXNIP-cg19693031 in peripheral blood was significantly associated with the development of incident T2D: after 8 years of follow-up, the relative risk of developing T2D was 1.09 (p value 1.2 × 10−17) per 1% decrease in methylation for TXNIP-cg19693031 (Chambers et al., 2015). Little is known about how this TXNIP DNAm site affects inflammation in relation to T2D. The present study investigated (1) the epigenetic association of TXNIP-cg19693031 with levels of selected inflammatory biomarkers and (2) whether the epigenetic association of TXNIP-cg19693031 with T2D is influenced by adjusting these inflammatory biomarkers in the model.
Materials and Methods
Study Sample
The Emory Twin Study consists of 283 middle-aged male Caucasian monozygotic (MZ) and dizygotic (DZ) twin pairs from the Vietnam Era Twin Registry (Goldberg et al., 2002) who were on active duty in the US military during the Vietnam conflict era (1964–1975; Vaccarino et al., 2008). Twins were examined in pairs at the Emory University General Clinical Research Center at baseline visits conducted between 2002 and 2010. Twins were given the same diet the evening before the assessments and instructed to refrain from smoking. All medications were held for approximately 24 h prior to testing. All measurements were performed in the morning after an overnight fast, and both members of each twin pair were examined at the same time. A medical history and a physical exam were obtained from all twins. Venous blood samples were drawn for the collection of plasma and peripheral blood leukocytes (PBL) and stored at −80 °C. Biochemical assays for each twin pair were processed in the same analytical run. Information on zygosity was determined by DNA analysis. A sub-group of 220 twins with sufficient amount of genomic DNA were successfully epityped using the Illumina HumanMethylation450 (450K) Beadchip. The Emory Twin Study was approved by the Emory Institutional Review Board, and all participants signed an informed consent.
Phenotypes
Weight and height were measured and used to calculate body mass index (BMI). Waist circumference was measured in centimeters at the level of the umbilicus. Cigarette smoking status was self-reported and dichotomized into current smokers versus noncurrent smokers (i.e. never or past smokers). As previously described, physical activity was categorized as ‘no physical activity’, ‘moderate physical activity’ and ‘vigorous physical activity’ (Vaccarino et al., 2014). Plasma levels of VCAM-1, ICAM-1, interleukin 6 (IL-6), soluble P-selectin, high-sensitivity c-reactive protein (hs-CRP), matrix metalloproteinase-2 (MMP-2), matrix metalloproteinase-9 (MMP-9), soluble RAGE (sRAGE) and fibrinogen were assessed using commercially available ELISA kits from R and D Systems (Minneapolis, Minnesota). Total triglycerides and cholesterol were determined by enzymatic methods (Beckman Coulter Diagnostics, Fullerton, CA). Levels of blood glucose, insulin and HbA1c in fasting plasma were measured on the Beckman CX7 chemistry auto analyzer (Beckman Coulter Diagnostics). Diabetes mellitus was defined as having a fasting glucose level of >126 mg/dl or being treated with antidiabetic medications.
DNA Methylation Profiling and Data Processing
As previously described, bisulfite convert genomic DNA samples from PBLs were whole-genome amplified, enzymatically fragmented and purified. Following standard protocols, samples were then hybridized to the 450K BeadChip, then fluorescently stained, scanned and assessed for fluorescence intensities at each bead site (Huang et al., 2018). Each DNAm site was quantified using probe intensities and beta (β) values, which represent the ratio of fluorescence intensity of the methylated and unmethylated sites (ranging from 0 to 1). Using the detection p-value threshold of .001, two individual samples with a missing rate above 5% were excluded, resulting in 218 eligible twins in the following analyses. We further performed data normalization and batch effects correction using the preprocessSWAN function of the ‘minfi’ package (Aryee et al., 2014) available in R.
Statistical Analysis
To compare the characteristics of participants with and without diabetes, generalized estimating equations (GEE) were applied to adjust for the relatedness between co-twins with T2D status as dichotomous dependent variable.
To understand the association between DNA methylation at TXNIP-cg19693031 (the DNAm site) and phenotypes, we applied two sets of regression analyses. We first modeled the fixed effects of twin-pair average methylation level at the DNAm site (shared epigenetic association) and individual deviation from the twin-pair average (i.e. the within twin-pair effect, representing the unshared epigenetic association in co-twins; Carlin et al., 2005). The second term also represents an association relating to individual specific factors that vary between co-twins (i.e. different lifestyles; Carlin et al., 2005). We then modeled the twins as individuals, accounting for within-twin correlation, to examine overall epigenetic associations with the outcomes.
For each set of analyses, we fitted 13 linear mixed regression models for twin study for four diabetes-related traits (HbA1c, fasting glucose, insulin and waist circumference) and nine inflammatory biomarkers as continuous dependent variables, respectively. To understand the association between DNA methylation and odds of T2D, we fitted one GEE model for T2D status as dichotomous dependent variable. Age in years, current smoking status and BMI were adjusted in all models as covariates. The linear mixed model contained random effects to address the chip/batch effect and the correlation within co-twins in linear mixed models, and GEE model addressed the correlation within co-twin by fitting exchangeable covariance matrix. DNAm was measured in PBL samples, where varying proportions of leukocyte subtypes and differential methylation in leukocyte subtypes may produce spurious epigenetic associations (Baron et al., 2006). We therefore adjusted for the proportions of PBL subtypes (B cells, granulocytes, monocytes, NK cells and T cells) in both models using the method developed by Houseman et al (2012).
To examine the effects of inflammatory biomarkers on the association between the DNAm site and T2D, we added them stepwise to a GEE model that accounted for correlations within twin pairs. We used stepwise, linear mixed models to examine the effects of inflammatory biomarkers on the associations between the DNAm site and diabetes-related traits (HbA1c, insulin, and glucose).
All analyses were conducted in R studio with R version 3.4.3. The R packages nlme and geepack were used to implement linear mixed effect models and GEE, respectively. To correct for multiple testing, we established a Bonferroni threshold for statistical significance (nominal p value < .004, correcting for 14 tested biomarkers).
Results
Table 1 summarizes the characteristics of 218 twin participants in the present study, including 164 monozygotic and 54 dizygotic twins (108 pairs and 2 singletons) stratified by the status of T2D. The mean age of the T2D group (n = 24) was 56.1 years old, similar to the non-T2D group (n = 194, mean age of 55.5). Compared to the non-T2D group, the T2D group had higher BMI and waist circumference, as well as higher levels of diabetes-related laboratory measures, including HbA1c, fasting glucose and insulin. Participants with T2D had a higher prevalence of coronary heart disease than did participants without T2D (33% versus 16%). A higher level of inflammation biomarkers was observed among patients with T2D, ICAM-1 (904 ng/ml versus 628 ng/ml), VCAM-1 (404 ng/ml versus 318 ng/ml) and MMP-2 (189 ng/ml versus 166ng/ml). As observed in previous studies (Chambers et al., 2015; Florath et al., 2016; Kulkarni et al., 2015; Mathur et ak., 2019; Meeks et al., 2019; Soriano-Tarraga et al., 2016; Walaszczyk et al., 2018), participants with T2D had lower DNAm level at TXNIP-cg19693031 (Table 1).
Table 1.
Characteristics of study participants, stratified by type 2 diabetes status (n = 218)
| Characteristics | T2D (n = 24) | Non-T2D (n = 194) |
p value |
|---|---|---|---|
| Age (yrs) | 56.1 (3.2) | 55.5 (3.3) | .52 |
| Body mass index (kg/m2) | 32.2 (3.79) | 29.0 (4.54) | .03 |
| Waist circumference (cm) | 108 (8.79) | 98.3 (11.6) | .001 |
| HbA1c (%) | 7.01 (1.29) | 5.49 (0.51) | <.001 |
| Insulin (mIU/l) | 21.1 (42.4) | 8.47 (6.97) | .05 |
| Glucose (mg/dL) | 132 (56.9) | 99.2 (13.0) | <.001 |
| ICAM-1 (ng/ml) | 904 (741) | 628 (231) | .003 |
| VCAM-1 (ng/ml) | 404 (254) | 318 (104) | <.001 |
| IL-6 (pg/ml) | 2.4 (2.17) | 2.09 (2.15) | .72 |
| P-selectin (ng/ml) | 98.5 (39.7) | 101 (29.6) | .81 |
| High-sensitivity CRP (mg/l) | 2.69 (2.65) | 2.95 (6.02) | .58 |
| MMP-2 (ng/ml) | 189 (61.9) | 166 (29.0) | .001 |
| MMP-9(ng/ml) | 337(157) | 334 (181) | .92 |
| sRAGE (pg/ml) | 996 (521) | 976 (435) | .86 |
| Fibrinogen (mg/dl) | 357 (77.5) | 351 (88.0) | .85 |
| DNAm level at TXNIP-cg19693031 | 0.69 (0.06) | 0.74 (0.05) | <.001 |
| Any medication for diabetes | 21 (0.88) | – | – |
| Current smoker | 8 (0.33) | 56 (0.29) | .48 |
| History of coronary heart disease | 8 (0.33) | 31 (0.16) | .10 |
| Hypertension | 8 (0.33) | 64 (0.33) | .44 |
| Physical activity (3 categories) | .71 | ||
| No physical activity | 2 (0.09) | 24 (0.23) | |
| Moderate physical activity | 15 (0.62) | 120 (0.62) | |
| Vigorous physical activity | 7 (0.29) | 50 (0.26) |
Note: T2D, type 2 diabetes. Data are shown as mean (SD) for continuous variables and as number of individuals (proportions) for categorical variables. p values are calculated from GEE models with occurrence of diabetes as the dependent variable. Bold type was used for significant p values.
T2D-related traits — including HbA1c, fasting glucose and insulin — were significantly associated with DNAm levels at TXNIP-cg19693031 after correcting for multiple testing and adjusting for age, BMI, current smoking status, proportions of PBL subtypes and technical batch effects (Table 2). We modeled the unshared association between co-twins by implementing the within-pair association; we also examined twins as individuals while accounting for co-twin correlations. Within twin pairs, 0.01 unit (1%) increase of DNAm beta values was associated with 0.0630 unit decrease in HbA1c (p value 9.84 × 10−4), as well as 24% lower prevalence of T2D (OR = 0.761, 95% CI [0.608, 0.953]). In the twin-as-individual analysis (which does not partition shared and unshared associations in co-twins), an 0.01 unit (1%) increase in DNAm beta value was independently associated with 0.05 unit decrease in HbA1c (p value 9.92 × 10−5), as well as 20% lower prevalence of T2D (OR = 0.802, 95% CI [0.708, 0.909]). Among nine tested inflammatory biomarkers, VCAM-1, ICAM-1, MMP-2 and P-selectin were significantly associated with DNAm levels at TXNIP-cg19693031 (Table 2). For both models, increase in DNAm beta value was associated with decrease in ICAM-1, VCAM-1 and MMP-2, while the VCAM-1 had the largest effect size. Within twin pairs, 0.01 unit increase in DNAm beta value was associated with 36.0 ng/ml decrease in VCAM-1 (p value of 2.78 × 10−4). Without partitioning the shared and unshared associations between co-twins (i.e. twin-as-individual analysis), a 1% increase in beta values was independently associated with 26.7 ng/ml decrease in VCAM-1 (p value of 4.33 × 10−7). The decrease in P-selectin was only associated with increase in DNAm in the first model. Since some inflammatory biomarkers have moderately skewed distribution, we also examined the log transformation of the analysis. The results of VCAM-1, ICAM-1 and MMP-2 were consistent (Supplementary Table S1). To further understand the effect of unshared environmental factors on DNA methylation, we repeated the analysis among a subset of monozygotic twins (Table 3) who shared the same genetic profile in addition to other demographic and familial factors. Comparing the results from all twins (Table 2), the significant associations in twin-specific models (i.e. HbA1c, insulin, ICAM-1 and P-selectin) and twins-as-individual models (i.e. HbA1c, insulin, glucose, ICAM-1, VCAM-1, MMP-2 and sRAGE) showed similar effect sizes (Table 3) with overlapping 95% confidence intervals. For instance, within twin pairs, a 0.01 unit increase in DNAm beta value was associated with 36.0 ng/ml decrease in ICAM-1 among all twins (95% CI −20.13, −7.47]) and the effect size was 9.27 ng/ml among monozygotic twins (95% CI [−14.9, −3.64]), while the effect size for twin-as-individual models was 8.02 ng/ml (95% CI [−11.69, −4.35]) and 7.05 ng/ml (95% CI [−10.6, −3.5]) among monozygotic twins. We also observed the same overlaps among the effect sizes of diabetes-relevant traits. Within twin pairs, a 0.01 unit (1%) increase of DNAm beta values was associated with 0.0630 unit decrease in HbA1c (95% CI [−0.1, −0.03]) and the effect size was 0.0843 units among monozygotic twins (95% CI [−0.13, −0.04]), while the effect size for twin-as-individual models was 0.0469 units (95% CI [−0.07, −0.02]) and 0.0568 units (95% CI [−0.08, −0.03]) among monozygotic twins.
Table 2.
Association of methylation at TXNIP-cg19693031 with diabetes-related and inflammatory biomarkers. The associations were examined in twin-specific models using individual deviation from the twin-pair average (within-pair effect) and twins-as-individual models
| Phenotype | N | Within-pair methylation | Individual methylation | ||||
|---|---|---|---|---|---|---|---|
| T2D-relevant traits | Beta | SE | p value | Beta | SE | p value | |
| HbA1c (%) | 215 | −0.0630 | 0.0185 | 9.84 × 10−4 | −0.0469 | 0.0115 | 9.92 × 10−5 |
| Insulin (mIU/L) | 217 | 0.873 | 0.492 | 0.08 | −0.567 | 0.234 | 0.02 |
| Glucose(mg/dL) | 217 | 0.574 | 0.640 | 0.37 | −1.11 | 0.375 | 0.004 |
| waist Circumference (cm) | 217 | −0.052 | 0.159 | 0.75 | 0.0150 | 0.0899 | 0.87 |
| Inflammatory biomarkers | p value* | p value* | |||||
| ICAM-1 (ng/ml) | 215 | −13.8 | 3.23 | 4.30 × 10−5 | −8.02 | 1.87 | 4.25 × 10−5 |
| VCAM-1 (ng/ml) | 215 | −36.0 | 9.55 | 2.78 × 10−4 | −26.7 | 4.92 | 4.33 × 10−7 |
| MMP-2 (ng/ml) | 215 | −2.97 | 0.870 | 9.52 × 10−4 | −2.47 | 0.528 | 9.23 × 10−6 |
| P-selectin (ng/ml) | 215 | −1.87 | 0.610 | 0.003 | −1.20 | 0.445 | 0.008 |
| IL-6 (pg/mL) | 212 | 0.0960 | 0.07 | 0.15 | 0.0132 | 0.0337 | 0.70 |
| High-sensitivity CRP (mg/L) | 214 | 0.179 | 0.182 | 0.33 | −0.00170 | 0.0883 | 0.99 |
| sRAGE (pg/mL) | 215 | −9.67 | 9.93 | 0.33 | −19.0 | 6.69 | 0.01 |
| Fibrinogen (mg/dL) | 198 | −1.96 | 2.31 | 0.34 | 0.672 | 1.41 | 0.63 |
| MMP-9 (ng/ml) | 215 | 3.54 | 5.31 | 0.51 | −2.31 | 2.77 | 0.41 |
Note: T2D, type 2 diabetes. *Bonferroni correction for multiple testing was used (p value <.005). Bold type was used for significant p values (<.05 for T2D-relevant traits and <.005 for inflammatory biomarkers). The analysis of each diabetes-related trait and inflammatory biomarker is adjusted for BMI, current smoking status, age and proportions of PBL subtypes (B cells, granulocytes, monocytes, NK cells and T cells). The table is sorted on within-pair comparison-associated p values in each category. The effect size is based on 0.01 unit (1%) difference in DNA methylation.
Table 3.
Association of methylation at TXNIP-cg19693031 with diabetes-related and inflammatory biomarkers among monozygotic twins. The associations were examined in twin-specific models using individual deviation from the twin-pair average (within-pair effect) and twins-as-individual models
| Phenotype | N | Within-pair methylation | Individual methylation | ||||
|---|---|---|---|---|---|---|---|
| T2D-relevant traits | Beta | SE | p value | Beta | SE | p value | |
| HbA1c (%) | 162 | −0.0843 | 0.0233 | 0.001 | −0.0568 | 0.0142 | 1.57 × 10−4 |
| Insulin (mIU/L) | 163 | 1.42 | 0.658 | 0.04 | −0.769 | 0.301 | 0.01 |
| Glucose(mg/dL) | 163 | 0.403 | 0.849 | 0.64 | −1.55 | 0.466 | 0.001 |
| waist Circumference (cm) | 163 | −0.188 | 0.183 | 0.31 | −0.0236 | 0.102 | 0.82 |
| Inflammatory biomarkers | p value* | p value* | |||||
| ICAM-1 (ng/ml) | 161 | −9.27 | 2.87 | 0.002 | −7.05 | 1.81 | 2.16 × 10−4 |
| VCAM-1 (ng/ml) | 161 | −29.3 | 10.7 | 0.008 | −26.6 | 5.15 | 2.13 × 10−6 |
| MMP-2 (ng/ml) | 161 | −2.49 | 1.01 | 0.02 | −2.47 | 0.584 | 7.14 × 10−5 |
| P-selectin (ng/ml) | 161 | −2.13 | 0.687 | 0.003 | −1.39 | 0.514 | 0.008 |
| IL-6 (pg/mL) | 158 | −0.0162 | 0.0579 | 0.78 | −0.0442 | 0.0314 | 0.16 |
| High-sensitivity CRP (mg/L) | 161 | −0.0606 | 0.147 | 0.68 | −0.0490 | 0.0634 | 0.44 |
| sRAGE (pg/mL) | 161 | −15.4 | 9.87 | 0.12 | −25.0 | 6.90 | 5.52 × 10−4 |
| Fibrinogen (mg/dL) | 152 | −3.11 | 2.53 | 0.22 | 0.114 | 1.58 | 0.94 |
| MMP-9 (ng/ml) | 161 | −5.22 | 4.59 | 0.26 | −5.63 | 2.71 | 0.04 |
Note: T2D, type 2 diabetes. *Bonferroni correction for multiple testing was used (p value < .005). Bold type was used for significant p values (<.05 for T2D-relevant traits and <.005 for inflammatory biomarkers). The analysis each diabetes-related trait and inflammatory biomarker is adjusted for BMI, current smoking status, age and proportions of PBL subtypes (B cells, granulocytes, monocytes, NK cells and T cells). Phenotypes are displayed in the same order as in the Table 2. The effect size is based on 0.01 unit (1%) difference in DNA methylation.
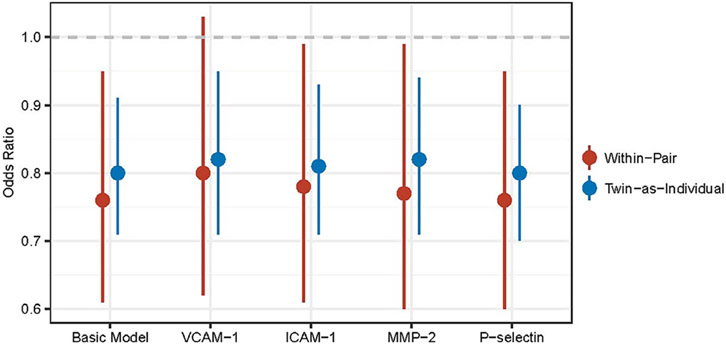
We then examined the effect of inflammatory biomarkers on the association between DNAm at TXNIP-cg19693031 and T2D using twin-specific GEE models for within-pair effects. Treating T2D as the outcome, the basic model included age, BMI, current smoking status and proportions of PBL subtypes as covariates. Each of the four inflammatory biomarkers significantly associated with DNAm at TXNIP-cg19693031(i.e. ICAM-1, VCAM-1, MMP-2 and P-selectin) was added to the basic model separately. The association of DNAm at TXNIP-cg19693031 with T2D was unchanged after adjusting for plasma levels of individual inflammatory biomarkers (Table 4, Figure 1). Both the unshared (within-pair) and overall (twin-as-individual) epigenetic associations remained significant, except in the model adjusting for plasma level of VCAM-1. VCAM-1 was independently associated with T2D (OR = 1.54, 95% CI [1.03, 2.31]), which slightly reduced the within-twin pair association to OR of 0.761 (95% CI [0.616, 1.03], p value .09). The analysis of diabetes-related traits (HbA1c, insulin and glucose) and TXNIP-cg19693031 showed consistent results (Table 4). The overall epigenetic associations of TXNIP-cg19693031 with HbA1c, insulin and glucose were all negative, indicating that lower levels of HbA1c and insulin and glucose were associated with increase in DNA methylation of TXNIP-cg19693031. The overall epigenetic associations of TXNIP-cg19693031 with these three traits remained significant after controlling for levels of inflammatory biomarkers.
Table 4.
Summary of twin-as-individual association of TXNIP-cg19693031 methylation with T2D, HbA1c, insulin and glucose adjusted for levels of individual inflammatory biomarkers
| Models | T2D | HbA1c | Insulin | Glucose | ||||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p value | Beta (95% CI) | p value | Beta (95% CI) | p value | Beta (95% CI) | p value | |
| Basic model | 0.802 (0.708, 0.909) | 5.25 × 10−4 | −0.0467 (−0.0698, −0.0235) | 1.27 × 10−4 | −0.571 (−1.03, −0.010) | 1.81 × 10−2 | −1.11 (−1.87, −0.36) | 4.27 × 10−3 |
| VCAM-1 | 0.824 (0.720, 0.954) | 9.42 × 10−3 | −0.0470 (−0.0718, −0.0223) | 2.79 × 10−4 | −0.633 (−1.16, −0.102) | 2.0 × 10−2 | −1.34 (−2.13, −0.53) | 1.5 × 10−3 |
| ICAM-1 | 0.815 (0.710, 0.934) | 2.99 × 10−3 | −0.0457 (−0.0699, −0.0214) | 3.27 × 10−4 | −0.575 (−1.06, −0.084) | 2.22 × 10−2 | −1.29 (−2.08, −0.50) | 1.59 × 10−3 |
| MMP-2 | 0.817 (0.707, 0.944) | 6.02 × 10−3 | −0.0468 (−0.0712, −0.0224) | 2.52 × 10−4 | −0.519 (−1.01, −0.030) | 3.78 × 10−2 | −1.24 (−2.04, −0.43) | 2.98 × 10−3 |
| P-selectin | 0.797 (0.702, 0.904) | 4.04 × 10−4 | −0.0476 (−0.0710, −0.0241) | 1.16 × 10−4 | −0.565 (−1.04, −0.090) | 2.02 × 10−2 | −1.15 (−1.91, −0.38) | 3.63 × 10−3 |
Note: T2D, type 2 diabetes; twin-as-individual, effect of overall measured influences; OR, odds ratio; beta, the coefficient estimates from the models; CI, confidence interval. In the basic model, BMI, current smoking status, age and proportions of PBL subtypes (B cells, granulocytes, monocytes, NK cells and T cells) were included as covariates, with T2D, HbA1c, insulin and glucose treated as the outcome, respectively. The other four models were constructed by adding corresponding inflammatory biomarkers one at a time into the basic model. Odds ratios of diabetes, beta of HbA1c, beta of insulin and beta of glucose are per 1% increase in DNA methylation and per 1 unit of standard deviation increase in inflammatory biomarkers.
Fig. 1.
Association of TXNIP-cg19693031 methylation with type 2 diabetes (T2D, adjusted for levels of individual inflammatory biomarkers). Odds ratio is represented by a dot; 95% CI is represented by each vertical line; lines that do not cross null effect (dashed line in grey) indicate statistically significant results; within-pair is the effect of nonshared measured influences; twin-as-individual is the effect of overall measured influences. In the basic model, BMI, current smoking status, age and proportions of PBL subtypes (B cells, granulocytes, monocytes, NK cells and T cells) were included as covariates, with T2D treated as the outcome. The other four models were constructed by adding inflammatory biomarkers one at a time into the basic model. Odds ratios for diabetes are per 1% increase in DNA methylation.
Discussion
Epigenetic variations can be influenced by genetic and environmental factors. Previous studies have examined epigenetic modification as a bridge between environmental exposures and human diseases. In the present analysis, we took advantage of a twin study population and a twin-specific statistical model to compare DNA methylation levels both between members of twin pairs and among all individuals in the study population. We used this approach to consider how DNA methylation changes might reflect both unshared and shared environmental or familial factors (Carlin et al., 2005).
This study showed that hypomethylation at TXNIP-cg19693031 is associated with higher odds of T2D and higher plasma levels of four inflammatory biomarkers: VCAM-1, ICAM-1, MMP-2 and P-selectin. Since some inflammatory biomarkers have moderately skewed distribution, we also examined the log transformation of the analysis. The result of VCAM-1, ICAM-1 and MMP-2 were consistent (Supplementary Table S1). The epigenetic associations we found for TXNIP-cg19693031 with T2D and related traits (HbA1c, fasting glucose and insulin) are consistent with previously reported findings (Chambers et al., 2015; Meeks et al., 2019). Our discovery of epigenetic associations of TXNIP-cg19693031 with inflammatory biomarkers suggests possible epigenetic linkage of TXNIP with endothelial cell activation via VCAM-1, ICAM-1 and P-selectin. We found, however, that levels of these inflammatory biomarkers did little to explain the epigenetic association between TXNIP-cg19693031 and T2D.
TXNIP, as a regulator of the cellular redox state, responds to high concentrations of glucose early in diabetes development. TXNIP transcription is largely induced by glucose via the activation of carbohydrate response element-binding protein (ChREBP), which binds to the TXNIP promotor (Cha-Molstad et al., 2009). Overexpression of TXNIP subsequently upregulates the oxidative stress response causing β-cell apoptosis (Minn et al., 2005). A previous study showed that cg19693031 is located in the 3’UTR of TXNIP, a site associated with glucose import and protein binding; methylation differences at this locus may affect the function of methylation-sensitive transcription regulators (Soriano-Tarraga et al., 2016). This is consistent with the association between T2D and hypomethylation at TXNIP-cg19693031 found in our study.
TXNIP also plays significant roles in inflammatory responses (L. Li et al., 2019; Nagai et al., 2007; Perrone et al., 2009; Szpigel et al., 2018; Wongeakin et al., 2014). In response to oxidative stress, TXNIP can bind to NOD-like receptor family, pyrin domain containing 3 (NLRP3) and activate the NLRP3-inflammasome to regulate inflammation in peripheral blood mononuclear cells (Szpigel et al., 2018) and in the cortex of postmortem human brain samples (L. Li et al., 2019). In addition, TXNIP has been found to induce NF-kB nuclear translocation and increase expression of inflammatory gene COX2 in rat retinal endothelial cells in culture (Perrone et al., 2009). NF-kB is also known to be involved in the diabetes-induced retinal expression of ICAM-1 and VEGF (Nagai et al., 2007). In the present study, we found that inflammatory biomarkers VCAM-1, ICAM-1, MMP-2, sRAGE and P-selectin were negatively associated with methylation at TXNIP-cg19693031. Some inflammatory biomarkers are related to endothelial dysfunction, which is considered a major cause of diabetes complications. Both ICAM-1 and VCAM-1 are pro-inflammatory markers expressed on endothelial cells and upregulated in peripheral blood during inflammation (X. Li et al., 2017). MMP-2 belongs to the matrix metalloproteinase family responsible for cellular proliferation, angiogenesis and inflammation (Fingleton, 2017). P-selectin initiates leukocyte recruitment corresponding to inflammatory action (Varga-Szabo et al., 2008). Previous studies on TXNIP and inflammatory biomarkers mainly focused on the protein level and RNA level using in vivo experimental design. Our study was the first to identify the epigenetic association between DNA methylation of TXNIP and inflammatory biomarkers.
In addition, we found similar results to previous research that found that T2D patients had a higher level of inflammatory biomarkers, including ICAM-1, VCAM-1 and MMP-2. Serum levels of CRP, IL-6, ICAM-1 were positively associated with T2D in previous studies (Gu et al., 2012; Hegazy et al., 2020; Vinagre et al., 2014). However, the association between MMPs and T2D is not consistent in the literature, showing a mixture of lower or higher concentration of MMPs in T2D patients (Lewandowski et al., 2011; Sampson et al., 2004), or no significant difference in MMPs concentration between T2D patients and controls (Papazafiropoulou et al., 2010). In the present study, MMP-9 showed no association with T2D while MMP-2 was positively associated with T2D. DNAm at TXNIP-cg19693031 is strongly associated with plasma levels of VCAM-1, ICAM-1, MMP-2 and P-selectin; however, we found that adjusting for these inflammatory biomarkers had little effect on the association of TXNIP-cg19693031 with T2D. Thus, the epigenetic association between TXNIP-cg19693031 and T2D is unlikely mediated via inflammatory response.
In the present study, the power and precision of estimates was limited by the small number of T2D cases. Homogeneity of the study cohort — all middle-aged male veterans — limits the generalizability of our findings to other populations with different characteristics. The present study used blood cells for DNAm profiling, which may not accurately represent the epigenetic modifications in other tissues directly related to T2D (e.g. pancreatic cells and adipose tissues) or inflammation (e.g. liver). Although studies of other disease-related tissues would be ideal, they may not be feasible. Large reference panels of epigenetic markers across many different tissues or cell types from the same individuals may help provide evidence for correlated epigenetic profiles across tissues and improve interpretability of blood-based epigenetic studies (Rivera & Ren, 2013). Finally, our data were cross-sectional and thus could not be used to evaluate epigenetic and phenotypic changes over time, including T2D incidence. The lack of longitudinal data limits our ability to establish causal relationships between DNAm, inflammation and T2D.
Epigenetics is an important molecular mechanism underlying many aspects of disease development and etiology. Initial EWAS of complex diseases, including T2D, have successfully identified strong epigenetic associations with relatively small sample sizes, compared with those required for genome-wide association studies. However, the molecular functions of the epigenetic markers identified by such studies are mostly unknown, limiting their potential contribution to understanding disease mechanisms and their utility for risk assessment and disease prevention. Therefore, it is important to conduct in-depth, follow-up studies of epigenetic markers to understand their biological roles. Our study of TXNIP-cg19693031 identified strong epigenetic associations with several inflammatory biomarkers related to endothelial dysfunction; however, these biomarkers did not explain the association of TXNIP-cg19693031 with T2D. Our study showed that epigenetic markers can be independently associated with different phenotypes. Such an example may inspire more sophisticated evaluation of multiple disease-related traits and risk factors in future epigenetic epidemiologic studies of complex disease, to fully understand the role of epigenetic regulations in human disease.
Supplementary Material
Acknowledgments.
The USA Department of Veterans Affairs has provided financial support for the development and maintenance of the Vietnam Era Twin (VET) Registry. Numerous organizations have provided invaluable assistance in the conduct of this study, including Department of Defense; National Personnel Records Center, National Archives and Records Administration; the Internal Revenue Service; National Institutes of Health; National Opinion Research Center; the National Research Council, National Academy of Sciences; the Institute for Survey Research, Temple University. Most importantly, the authors gratefully acknowledge the continued cooperation and participation of the members of the VET Registry and their families. Without their contribution, this research would not have been possible.
Financial Support.
This work was partly supported by the National Institutes of Health grants NR013520, HL077506, HL68630, AG026255, MH056120, HL088726, NR013520 and MH076955; by the Emory University General Clinical Research Center MO1-RR00039; and by American Heart Association grants 0245115N and 13GRNT17060002.
Footnotes
Supplementary Material. To view supplementary material for this article, please visit https://doi.org/10.1017/thg.2021.42.
Ethical Standards. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
Conflict of Interest. None.
References
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, & Irizarry RA (2014). Minfi: A flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics, 30, 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron U, Turbachova I, Hellwag A, Eckhardt F, Berlin K, Hoffmuller U, Gardina P, & Olek S (2006). DNA methylation analysis as a tool for cell typing. Epigenetics, 1, 55–60. [DOI] [PubMed] [Google Scholar]
- Carlin JB, Gurrin LC, Sterne JA, Morley R, & Dwyer T (2005). Regression models for twin studies: A critical review. International Journal of Epidemiology, 34, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Cha-Molstad H, Saxena G, Chen J, & Shalev A (2009). Glucose-stimulated expression of Txnip is mediated by carbohydrate response element-binding protein, p300, and histone H4 acetylation in pancreatic beta cells. The Journal of Biological Chemistry, 284, 16898–16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JC, Loh M, Lehne B, Drong A, Kriebel J, Motta V, Wahl S, Elliott HR, Rota F, Scott WR, Zhang W, Tan ST, Campanella G, Chadeau-Hyam M, Yengo L, Richmond RC, Adamowicz-Brice M, Afzal U, Bozaoglu K, – Kooner JS (2015). Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: A nested case-control study. The Lancet. Diabetes & Endocrinology, 3, 526–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Saxena G, Mungrue IN, Lusis AJ, & Shalev A (2008). Thioredoxin-interacting protein: A critical link between glucose toxicity and beta-cell apoptosis. Diabetes, 57, 938–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingleton B. (2017). Matrix metalloproteinases as regulators of inflammatory processes. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research, 1864, 2036–2042. [DOI] [PubMed] [Google Scholar]
- Florath I, Butterbach K, Heiss J, Bewerunge-Hudler M, Zhang Y, Schottker B, & Brenner H (2016). Type 2 diabetes and leucocyte DNA methylation: an epigenome-wide association study in over 1,500 older adults. Diabetologia, 59, 130–138. [DOI] [PubMed] [Google Scholar]
- Goldberg J, Curran B, Vitek ME, Henderson WG, & Boyko EJ (2002). The Vietnam Era Twin Registry. Twin Research, 5, 476–481. [DOI] [PubMed] [Google Scholar]
- Gu HF, Ma J, Gu KT, & Brismar K (2012). Association of intercellular adhesion molecule 1 (ICAM1) with diabetes and diabetic nephropathy. Frontiers in Endocrinology (Lausanne), 3, 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegazy GA, Awan Z, Hashem E, Al-Ama N, & Abunaji AB (2020). Levels of soluble cell adhesion molecules in type 2 diabetes mellitus patients with macrovascular complications. Journal of International Medical Research, 48, 300060519893858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, & Kelsey KT (2012). DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics, 13, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Hui Q, Walker DI, Uppal K, Goldberg J, Jones DP, Vaccarino V, & Sun YV (2018). Untargeted metabolomics reveals multiple metabolites influencing smoking-related DNA methylation. Epigenomics, 10, 379–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui STY, Andres AM, Miller AK, Spann NJ, Potter DW, Post NM, Chen AZ, Sachithanantham S, Jung DY, Kim JK, & Davis RA (2008). Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proceedings of the National Academy of Sciences of the United States of America, 105, 3921–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodneva Y, Shalev A, Frank SJ, Carson AP, & Safford MM (2016). Calcium channel blocker use is associated with lower fasting serum glucose among adults with diabetes from the REGARDS study. Diabetes Research and Clinical Practice, 115, 115–121. doi: 10.1016/j.diabres.2016.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni H, Kos MZ, Neary J, Dyer TD, Kent JW Jr., Goring HH, Cole SA, Comuzzie AG, Almasy L, Mahaney MC, Curran JE, Blangero J, & Carless MA (2015). Novel epigenetic determinants of type 2 diabetes in Mexican-American families. Human Molecular Genetics, 24, 5330–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, & Mittal R (2018). Mapping Txnip: Key connexions in progression of diabetic nephropathy. Pharmacological Reports, 70, 614–622. [DOI] [PubMed] [Google Scholar]
- Lewandowski KC, Banach E, Bieńkiewicz M, & Lewiński A (2011). Matrix metalloproteinases in type 2 diabetes and non-diabetic controls: Effects of short-term and chronic hyperglycaemia. Archives of Medical Science, 7, 294–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ismael S, Nasoohi S, Sakata K, Liao F-F, McDonald MP, & Ishrat T (2019). Thioredoxin-interacting protein (TXNIP) associated NLRP3 inflammasome activation in human Alzheimer’s disease brain. Journal of Alzheimer’s Disease, 68, 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kover KL, Heruth DP, Watkins DJ, Guo Y, Moore WV, He LG, Zang M, Clements MA, & Yan Y (2017). Thioredoxin-interacting protein promotes high-glucose-induced macrovascular endothelial dysfunction. Biochemical and Biophysical Research Communications, 493, 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R, Hui Q, Huang Y, Gwinn M, So-Armah K, Freiberg MS, Justice AC, Xu K, Marconi VC, & Sun YV (2019). DNA methylation markers of type 2 diabetes mellitus among male veterans with or without human immunodeficiency virus infection. Journal of Infectious Diseases, 219, 1959–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks KAC, Henneman P, Venema A, Addo J, Bahendeka S, Burr T, Danquah I, Galbete C, Mannens MMAM, Mockenhaupt FP, Owusu-Dabo E, Rotimi CN, Schulze MB, Smeeth L, Spranger J, Zafarmand MH, Adeyemo A, & Agyemang C (2019). Epigenome-wide association study in whole blood on type 2 diabetes among sub-Saharan African individuals: Findings from the RODAM study. International Journal of Epidemiology, 48, 58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AH, Hafele C, & Shalev A (2005). Thioredoxin-interacting protein is stimulated by glucose through a carbohydrate response element and induces beta-cell apoptosis. Endocrinology, 146, 2397–2405. [DOI] [PubMed] [Google Scholar]
- Mortuza R, Chen S, Feng B, Sen S, & Chakrabarti S (2013). High glucose induced alteration of SIRTs in endothelial cells causes rapid aging in a p300 and FOXO regulated pathway. PLoS One, 8, e54514–e54514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai N, Izumi-Nagai K, Oike Y, Koto T, Satofuka S, Ozawa Y, Yamashiro K, Inoue M, Tsubota K, Umezawa K, & Ishida S (2007). Suppression of diabetes-induced retinal inflammation by blocking the angiotensin II type 1 receptor or its downstream nuclear factor-kappaB pathway. Investigative Ophthalmology & Visual Science, 48, 4342–4350. [DOI] [PubMed] [Google Scholar]
- Papazafiropoulou A, Perrea D, Moyssakis I, Kokkinos A, Katsilambros N, & Tentolouris N (2010). Plasma levels of MMP-2, MMP-9 and TIMP-1 are not associated with arterial stiffness in subjects with type 2 diabetes mellitus. Journal of Diabetes and Its Complication, 24, 20–27. [DOI] [PubMed] [Google Scholar]
- Perrone L, Devi TS, Hosoya K, Terasaki T, & Singh LP (2009). Thioredoxin interacting protein (TXNIP) induces inflammation through chromatin modification in retinal capillary endothelial cells under diabetic conditions. Journal of Cellular Physiology, 221, 262–272. [DOI] [PubMed] [Google Scholar]
- Rivera CM, & Ren B (2013). Mapping human epigenomes. Cell, 155, 39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson M, Davies I, Gavrilovic J, Sussams B, Brown J, Astley S, & Hughes DA (2004). Plasma matrix metalloproteinases, low density lipoprotein oxidisability and soluble adhesion molecules after a glucose load in Type 2 diabetes. Cardiovascular Diabetology, 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh LP, Devi TS, & Yumnamcha T (2017). The role of Txnip in mitophagy dysregulation and inflammasome activation in diabetic retinopathy: A new perspective. JOJ Ophthalmology, 4. doi: 10.19080/jojo.2017.04.555643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano-Tarraga C, Jimenez-Conde J, Giralt-Steinhauer E, Mola-Caminal M, Vivanco-Hidalgo RM, Ois A, Rodríguez-Campello A, Cuadrado-Godia E, Sayols-Baixeras S, Elosua R, & Roquer J (2016). Epigenome-wide association study identifies TXNIP gene associated with type 2 diabetes mellitus and sustained hyperglycemia. Human Molecular Genetics, 25, 609–619. [DOI] [PubMed] [Google Scholar]
- Szpigel A, Hainault I, Carlier A, Venteclef N, Batto AF, Hajduch E, Bernard C, Ktorza A, Gautier J-F, Ferré P, Bourron O, & Foufelle F (2018). Lipid environment induces ER stress, TXNIP expression and inflammation in immune cells of individuals with type 2 diabetes. Diabetologia, 61, 399–412. [DOI] [PubMed] [Google Scholar]
- Vaccarino V, Goldberg J, Magruder KM, Forsberg CW, Friedman MJ, Litz BT, Heagerty PJ, Huang GD, Gleason TC, & Smith NL (2014). Posttraumatic stress disorder and incidence of type-2 diabetes: A prospective twin study. Journal of Psychiatric Research, 56, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarino V, Lampert R, Bremner JD, Lee F, Su S, Maisano C, Murrah NV, Jones L, Farhan J, Nadeem A, Ashraf A, & Goldberg J (2008). Depressive symptoms and heart rate variability: evidence for a shared genetic substrate in a study of twins. Psychosomatic Medicine, 70, 628–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga-Szabo D, Pleines I, & Nieswandt B (2008). Cell adhesion mechanisms in platelets. Arteriosclerosis, Thrombosis, and Vascular Biology, 28, 403–412. [DOI] [PubMed] [Google Scholar]
- Vinagre I, Sánchez-Quesada JL, Sánchez-Hernández J, Santos D, Ordoñez-Llanos J, De Leiva A, & Pérez A (2014). Inflammatory biomarkers in type 2 diabetic patients: effect of glycemic control and impact of LDL subfraction phenotype. Cardiovascular Diabetology 13, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walaszczyk E, Luijten M, Spijkerman AMW, Bonder MJ, Lutgers HL, Snieder H, Wolffenbuttel BHR, & van Vliet-Ostaptchouk JV (2018). DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA(1c) levels: A systematic review and replication in a case-control sample of the Lifelines study. Diabetologia, 61, 354–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongeakin N, Bhattarakosol P, & Patumraj S (2014). Molecular mechanisms of curcumin on diabetes-induced endothelial dysfunctions: Txnip, ICAM-1, and NOX2 expressions. BioMed Research International, 2014, Article ID 161346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Chen J, Jing G, & Shalev A (2012). Preventing β-cell loss and diabetes with calcium channel blockers. Diabetes, 61, 848–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T, Kuo SC, Chang YY, Chen YT, & Wang KK (2017). Verapamil use is associated with reduction of newly diagnosed diabetes mellitus. Journal of Clinical Endocrinology and Metabolism, 102, 2604–2610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.