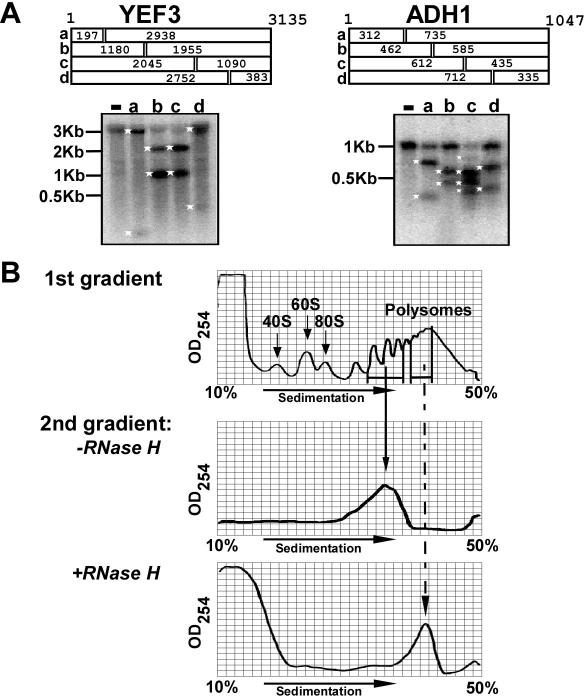
Figure 3.
Controls for RDM. (A) Specificity of the RNase H reactions. Antisense ODNs complementary to various positions of ADH1 or YEF3 were annealed to polysomal mRNA. The expected lengths (in nt) of the cleavage products are shown schematically at the top of each panel. Following annealing, RNase H was added and reactions proceeded for 20 min. Samples were then subjected to northern analysis using radiolabeled full-length DNA probe. Size markers are shown at the left and white asterisks indicate cleavage products. (−) No oligo added and the letters correspond to the schematic above. (B) No detectable ribosome dissociation during processing steps. The upper panel (first gradient) is an OD254 trace of a sucrose gradient from which two different polysomal fractions were collected. One fraction (mRNAs associated with 3–5 ribosomes) was resolved on a second gradient following incubation for 1 h at 37°C (second gradient, −RNase H panel) and the other fraction (mRNAs associated with 5–10 ribosomes) was annealed with an antisense ODN complementary to ADH1 around position 462, subjected to RNase H reaction, and then separated on a sucrose gradient (second gradient, +RNase H). The sedimentation positions of the 40S, 60S, 80S and polysomes are indicated.