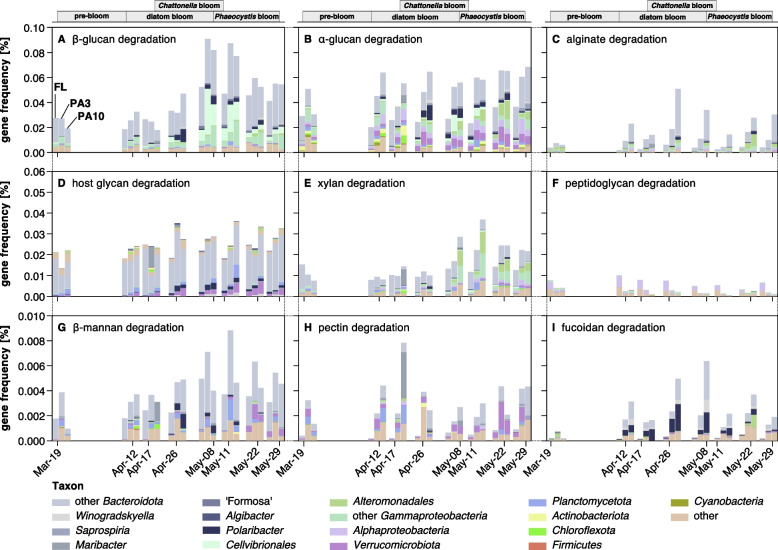
Fig. 4.
Frequencies of CAZyme genes attributed to the degradation of specific polysaccharide substrates. CAZyme genes in metagenomes and corresponding substrates were predicted using the dbCAN3-sub database. Gene frequencies were calculated as follows: frequency = Σ(average coverage of target genes) × 100/Σ(average coverage of all genes). Only dates with data for all fractions were plotted as stacked bar charts (left to right: FL, 0.2–3 µm; PA3, 3–10 µm; PA10, > 10 µm). Colors represent dominating taxa. Additional data for α-rhamnosides, chitin, arabinans, α-mannans, cellulose, and sialic acids is depicted in Additional file 2: Fig. S10, and complete data is summarized in Additional file 1: Table S7