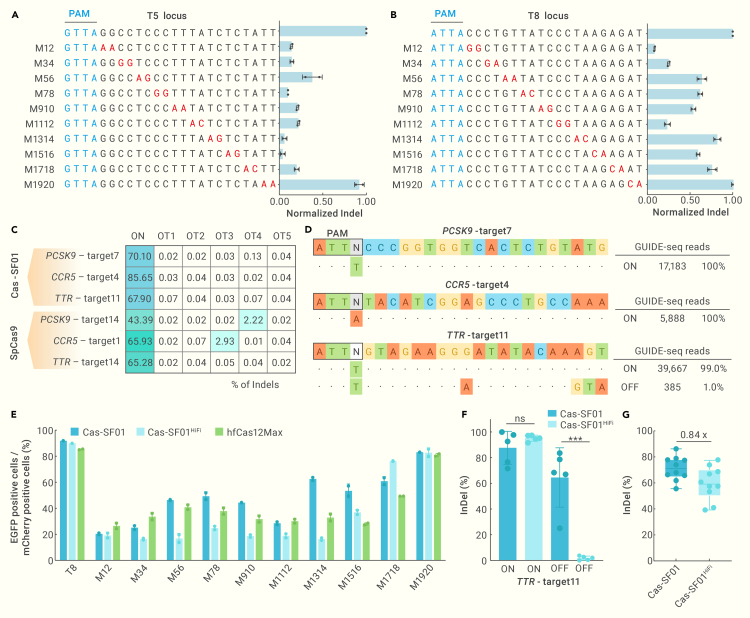
Figure 4.
Analysis of the specificity of Cas-SF01 and Cas-SF01HiFi
(A and B) The mismatch tolerance of Cas-SF01 was examined by designing dinucleotide mutations at T5 (A) and T8 (B) targets, and then the activation of EGFP was determined.
(C) Off-target rates of Cas-SF01 and SpCas9 at in silico predicted off-target sites, as determined by targeted deep sequencing.
(D) The genome-wide off-target effects of Cas-SF01 as analyzed by GUIDE-seq. The numbers of on-target and off-target reads are shown at right.
(E) The mismatch tolerance of Cas-SF01, Cas-SF01HiFi, and hfCas12Max at the T8 locus.
(F) The indel activities of Cas-SF01 and Cas-SF01HiFi at the on-target site TTR-target11 (ON) and off-target site (OFF) shown in (D). ∗∗∗p < 0.001; ns, not significant (Student’s t test).
(G) Comparison of indel activity of Cas-SF01 with Cas-SF01HiFi at 10 target sites at CCR5 locus. Each dot represents 2 replicates.