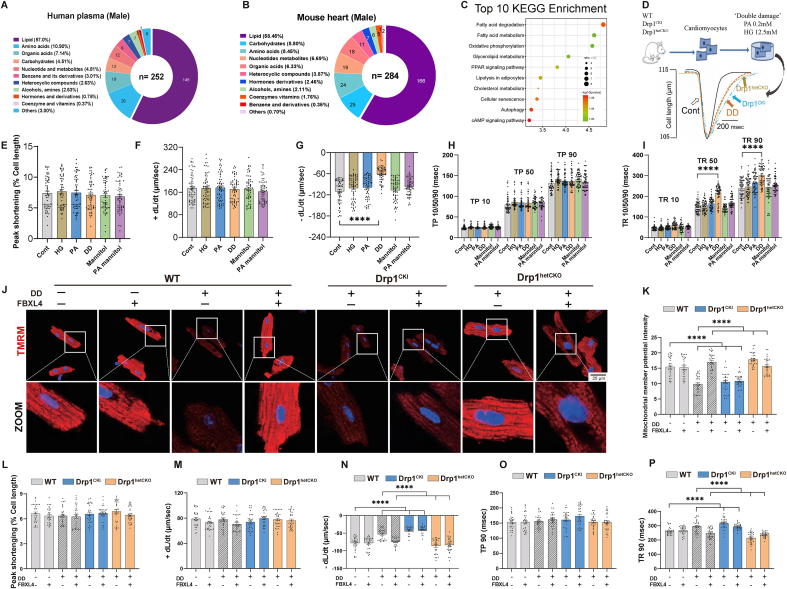
Fig. 6.
Metabolomics and proteomics analyses in plasma from HFpEF patients and mouse hearts, as well as establishment of an in vitro HFpEF model for diastolic dysfunction. A: Metabolomics analysis of human plasma (male only) using Mass spectrometry; B: Metabolomics analysis of mouse heart tissues; C: KEGG pathways of top 10 differential proteins from HFpEF mouse hearts using TMT-proteomics; D: Protocol depicting the in vitro ‘double-damage’ (DD) model established using high glucose (HG, 12.5 mM) and palmitic acid (PA, 0.2 mM) in cardiomyocytes from WT, Drp1CKI and Drp1hetCKO mice; E: Peak shortening (% cell length); F: Maximal velocity of shortening (+dL/dt); G: Maximal velocity of relengthening (-dL/dt); H: Time-to-10%, 50% and 90% shortening (TP10, TP50 and TP90); I: Time-to-10%, 50%, and 90% relengthening (TR10, TR50 and TR90); J: Representative tetramethylrhodamine, methyl ester (TMRM) staining of mitochondrial membrane potential (red color) using confocal microscopy; K: Pooled data of mitochondrial membrane potential using TMRM; L–P: Adult mouse cardiomyocytes from Drp1CKI and Drp1hetCKO mice were transfected with Adv-FBXL4 or vector for 48 h prior to ‘double-damage’ challenge for another 4 h. L: Peak shortening; M: + dL/dt; N: dL/dt; O: Time-to-90% shortening (TP90); and P: Time-to-90% relengthening (TR90). Mean ± SEM, Independent-samples used t-test or Wilcoxon rank sum test for continuous variables and Chi-square test for categorical variables (details provided in Supplemental Table 1); n = 15 subjects (panel A) for metabolomics per group; n = 6 mice (panel B) for metabolomics; n = 3 mice (panel C) for TMT-proteomics, Differential proteins were identified using variable important in projection (VIP) values of >1.2 in an orthogonal partial least square–discriminant analysis (OPLS-DA) model. n = 6 mice per group (10 replicates per heart, panel E–I), and n = 6 mice per group (5 replicates per heart, panel J–P). Statistical significance was estimated using one-way ANOVA followed by the Tukey's multiple comparison test, repeated measures ANOVA in R and Bonferroni adjusted test. ***p < 0.001, ****p < 0.0001 between indicated groups.