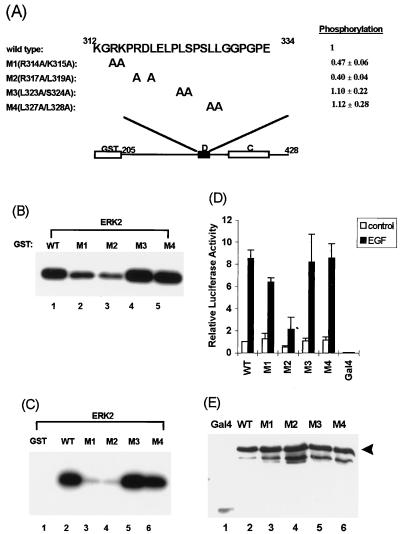
FIG. 7.
Mapping the D domain residues required for targeting of ERK2. (A) Amino acid sequences of the WT and D-domain mutants R314A/K315A (M1), R317A/L319A (M2), L323A/S324A (M3), and L327A/L328A (M4). Numbers above the sequence represent the N- and C-terminal residues in the D domain. The degree of phosphorylation of each protein (relative to WT Elk-205) is indicated. Standard deviations of the data from four independent experiments are indicated. (B) Immune-complex kinase assays of mutant GST–Elk-205 fusions by ERK2. The phosphorylation of GST–Elk-205 fusion proteins by ERK2 MAP kinase was examined by the immune-complex protein kinase assay. Kinase assays were performed for 15 min at 30°C with 2 U of activated ERK2 (NEB) and equal molar quantities (5 pmol) of GST–Elk-1 fusion proteins as substrates. (C) Binding and phosphorylation assays of wild-type and mutant GST-Elk proteins. Equal molar quantities of GST–Elk-205 fusion proteins were bound to ERK2 (50 U; NEB) and washed, and the remaining bound kinases were assayed by incubating with [γ-32P]ATP in kinase buffer for 2 h at 30°C. (D) EGF-stimulated transcriptional activation by wild-type and mutant GAL4–Elk-1 fusion proteins. COS-1 cells were transfected with cytomegalovirus promoter-driven constructs encoding GAL4 fusions to either WT or D-domain mutant Elk-1 derivatives and a GAL4-driven luciferase reporter plasmid. The cells were either unstimulated or stimulated with EGF. Transfection efficiency was monitored by using the β-galactosidase expression vector pCH110. The luciferase activities relative to GAL4–Elk-205-mediated reporter activation in unstimulated cells (means ± standard deviations; n = 3) are presented. (E) Expression levels of the GAL4 fusion proteins in COS-1 cells were examined with total-cell extracts for Western blot analysis with an anti-GAL4 antibody. Bands representing full-length GAL4–Elk-205 are indicated by the arrowhead.