ABSTRACT
In pandemic scenarios involving novel human pathogenic viruses, it is highly desirable that vaccines induce strong neutralizing antibodies as quickly as possible. However, current vaccine strategies require multiple immunization doses to produce high titers of neutralizing antibodies and are poorly protective after a single vaccination. We therefore wished to design a vaccine candidate that would induce increased protective immune responses following the first vaccine dose. We hypothesized that antibodies against the receptor-binding domain (RBD) of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike glycoprotein could be increased by drawing upon immunity to a previous infection. We generated a fusion protein containing the influenza H1N1 PR8 virus nucleoprotein (NP) and the SARS-CoV-2 spike RBD. Mice with or without preexisting immunity to PR8 were then vaccinated with NP/RBD. We observed significantly increased SARS-CoV-2 neutralizing antibodies in mice with PR8 immunity compared to mice without preexisting PR8 immunity. Vaccination with NP/RBD protected mice from SARS-CoV-2-induced morbidity and mortality after a single dose. Additionally, we compared SARS-CoV-2 virus titers in the lungs and nasal turbinates 4 days post-challenge of mice vaccinated with NP/RBD. SARS-CoV-2 virus was detectable in the lungs and nasal turbinate of mice without preexisting PR8 immunity, while SARS-CoV-2 virus was completely undetectable in mice with preexisting PR8 immunity. We also found that CD4-positive T cells in mice with preexisting immunity to PR8 play an essential role in producing the increased antibody response against RBD. This vaccine strategy potentially can be modified to target other pathogens of concern and offers extra value in future pandemic scenarios.
IMPORTANCE
Increased globalization and changes in human interactions with wild animals has increased the likelihood of the emergence of novel viruses with pandemic potential. Vaccines can be effective in preventing severe disease caused by pandemic viruses. However, it takes time to develop protective immunity via prime-boost vaccination. More effective vaccine designs should quickly induce protective immunity. We propose leveraging preexisting immunity to a different pathogen to boost protection against emerging viruses. We targeted SARS-CoV-2 as a representative pandemic virus and generated a fusion protein vaccine that combines the nucleoprotein from influenza A virus and the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein. Our vaccine design significantly increased the production of RBD-specific antibodies in mice that had previously been exposed to influenza virus, compared to those without previous exposure. This enhanced immunity reduced SARS-CoV-2 replication in mice. Our results offer a vaccine design that could be valuable in a future pandemic setting.
KEYWORDS: virology, vaccine, emerging virus, SARS-CoV-2
INTRODUCTION
Since the start of the 21st century, the world has seen the emergence of three novel human coronaviruses. Severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) was first identified in China in November 2002 (1), and Middle East respiratory syndrome coronavirus was identified in Saudi Arabia in June of 2012 (2). Then, at the end of 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified as a novel human coronavirus in China (3). The rapid spread of SARS-CoV-2 quickly caused a global pandemic on a scale not seen since the 1918 influenza pandemic. The rollout of effective vaccines against the virus has greatly reduced the overall burden of coronavirus disease 2019 (COVID-19) morbidity and mortality (4–6). However, the emergence of more transmissible virus variants capable of evading vaccine-derived immune responses has driven new surges of disease (7–12). It is therefore likely that SARS-CoV-2 infections will continue for some time, and vaccines will continue to be needed to combat the virus. Additionally, the emergence of three novel human coronaviruses in less than two decades makes it likely that another novel human coronavirus will emerge in the future. Changes in the global landscape that include increased globalization, rapid deforestation, and wildlife trafficking increase the likelihood of the emergence of zoonotic viruses with pandemic potential (13, 14). Vaccine strategies for future pandemic scenarios are, therefore, an important area of research and development.
The main vaccine strategies developed during the SARS-CoV-2 pandemic target the SARS-CoV-2 spike glycoprotein. Spike is the main surface viral antigen of SARS-CoV-2 that exists as a trimer on the surface of the virus particle, with each monomer consisting of a protein of approximately 180 kD in size made up of S1 and S2 subunits (15–17). During virus infection, the spike binds to angiotensin-converting enzyme 2 (ACE2) receptors on the surface of host cells. Binding to ACE2 triggers internalization of the virus into the cell and also results in conformational changes in the spike protein to allow for the fusion of the virus envelope with the host membrane, allowing for the release of the virus genome into the cytoplasm. Within the S1 subunit of the spike protein is the receptor-binding domain (RBD), which contains the contact points for spike binding to ACE2 (18). Vaccines targeting the RBD domain produce RBD-specific antibodies in mice, nonhuman primates, and humans. These RBD-specific antibodies can neutralize SARS-CoV-2 viruses, and RBD vaccines protect against COVID-19 in disease models and in humans (19–30).
While the vaccine strategies against SARS-CoV-2 have been remarkably effective at saving lives and mitigating severe disease, it takes weeks and multiple vaccination doses to achieve favorable antibody responses. Neutralizing antibodies against the USA-WA1/2020 (WA1) strain of SARS-CoV-2 were detected in only 79% of individuals vaccinated with the Moderna mRNA-1273 vaccine at 4 weeks after a single dose, compared to 100% of individuals 2 weeks after a second dose (31). Similarly, neutralizing antibody titers against SARS-CoV-2 were detected in only 73% of individuals vaccinated with 30 µg of BNT162b1 at 3 weeks after a single dose, compared with 100% of individuals 2 weeks after a second dose (5), and this difference was also observed with the BNT162b2 vaccine (32). In a study of nonhuman primates vaccinated with the mRNA-1273 vaccine, neutralization assays with a pseudotyped lentivirus reporter showed 50% inhibitory dilution (ID50) geometric mean titers (GMTs) of 63 at 4 weeks after the first vaccination, compared with GMT of 103 by 4 weeks after the second vaccination (33).
These lower antibody responses are associated with lower protection against disease. A study in Qatar found 83% effectiveness of the mRNA-1273 vaccine in 2021 against severe disease and death among individuals receiving only one dose of vaccine, compared to 97% vaccine effectiveness among individuals receiving two doses of vaccine (34). A study of vaccine effectiveness against emerging variants found that vaccine effectiveness against symptomatic disease following infection with the Delta variant was only 30.7% among individuals who received a single dose of the BNT162b2 vaccine while vaccine effectiveness was 79.6% among individuals who received two doses. This study also observed only 30.0% vaccine effectiveness among individuals who received a single dose of the ChAdOx1 vaccine, while vaccine effectiveness was 67% among individuals who received a second dose of the ChAdOx1 vaccine (35). Taken together, these studies point to an unmet need for vaccines against pandemic viruses that better protect people after a single vaccine dose.
DiPiazza et al. (36) have previously hypothesized that helper T-cell availability is a limiting factor in the production of robust antibody responses to novel avian influenza viruses. To overcome this limitation, they demonstrated that preexisting helper T-cell immunity to seasonal influenza viruses can be harnessed to increase antibody responses to novel avian influenza viruses (36). We similarly hypothesized that the limited availability of spike RBD-specific helper T cells could be overcome by harnessing preexisting immunity to a separate virus to increase antibody responses against SARS-CoV-2 spike protein. As a proof of concept, we tested whether antibody responses to the RBD domain of SARS-CoV-2 spike protein could be enhanced through vaccinating animals with preexisting immunity to influenza with a fusion protein containing influenza nucleoprotein (NP) and SARS-CoV-2 RBD protein (NP/RBD). Influenza virus was selected because most people have encountered influenza virus antigens. In this vaccine scenario, RBD-specific B cells are likely to obtain help from a population of previously expanded NP-specific helper T cells since RBD-specific B cells would present both NP and RBD antigenic peptides in their major histocompatibility complex (MHC) class II. Additionally, we theorized that NP-specific antibodies from a previous immunization could facilitate the Fc receptor-mediated uptake of NP/RBD by antigen-presenting cells (APCs), which would accelerate the presentation of antigen to RBD-specific helper T cells.
In our vaccination studies, we observed that vaccination with NP/RBD produced higher RBD-specific antibodies in mice with previous immunity to influenza compared to mice without previous immunity to influenza 14 days after vaccination. These results were observed when previous immunity to the influenza virus was established either through infection with a live virus or vaccination with an inactivated virus. Additionally, NP/RBD vaccination in the presence of immunity to influenza produced complete protection against weight loss and death in mice challenged with SARS-CoV-2. In these same challenged mice, SARS-CoV-2 virus was detectable in the lungs and nasal turbinate of mice without preexisting PR8 immunity, while SARS-CoV-2 virus was completely undetectable in mice with preexisting PR8 immunity. Furthermore, we demonstrated that CD4-positive T cells in mice with preexisting immunity to PR8 were necessary for the increased RBD-specific antibody response after NP/RBD vaccination. These results represent an improved vaccine strategy for future pandemic scenarios.
RESULTS
Purification of NP/RBD fusion protein and predicted structure
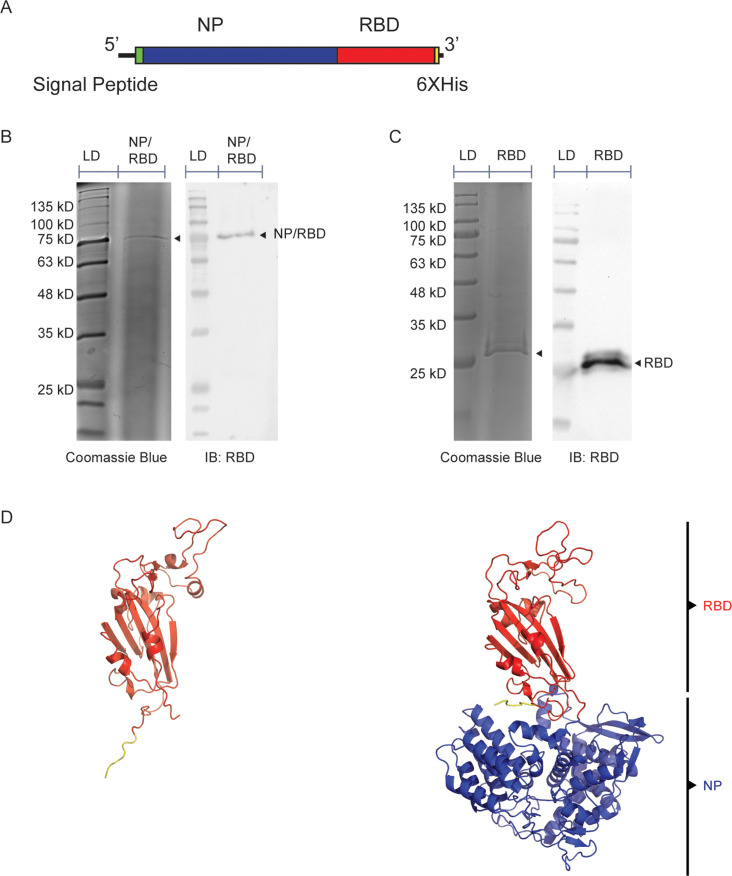
We used the baculovirus/SF9 insect cell expression system to express our chimeric NP/RBD fusion protein. The gene for the chimeric protein was cloned into the pFastBac plasmid for baculovirus rescue and contained an N-terminal secretion signal from the gene for the hemagglutinin protein of influenza virus A/WSN/33 H1N1 (WSN) strain. Influenza virus A/Puerto Rico/8/34 H1N1 (PR8) NP gene was included adjacent to the C-terminal end of the signal peptide and was fused to the RBD domain of the WA1 strain of SARS-CoV-2 (amino acids 330–528). A 6xHis tag was included at the C-terminal end (Fig. 1A). NP/RBD fusion protein was purified with a HisTrap Nickel column. The purity of the purified protein was verified by Coomassie blue stain, and its identity was confirmed by Western blot using an RBD-specific antibody (Fig. 1B). Unlinked RBD protein not fused to NP was also purified with a HisTrap column and verified by Coomassie blue staining and Western blot (Fig. 1C). Additionally, size exclusion chromatography of the purified NP/RBD protein was performed to increase the purity of the protein. However, the resulting fractions from the size exclusion chromatography were all positive for NP/RBD by Western blot (Fig. S1), suggesting that the NP/RBD fusion protein may form oligomers similar to unlinked NP protein (37). To test that the original conformations of NP and RBD proteins were maintained in the fusion protein, we used the AlphaFold program to examine the structure of the NP/RBD fusion protein (Fig. 1D) (38). The predicted protein structure showed NP and RBD folding separately, with RBD maintaining its original shape. After confirming the purity of the purified NP/RBD fusion protein, we proceeded to test its immunogenicity in a mouse model.
Fig 1.
Purification and structure prediction of NP/RBD protein and RBD protein. (A) Diagram of genetic structure of NP/RBD 6xHis. (B) Western blot and Coomassie blue stain of purified NP/RBD 6xHis (LD = protein ladder). (C) Western blot and Coomassie blue stain of purified RBD 6xHis (amino acids 330–528). (D) AlphaFold predicted structure of RBD 6xHis (amino acids 330–528) and NP/RBD 6xHis. RBD is shown in blue, 6xHis tag is shown in yellow, and NP is shown in orange.
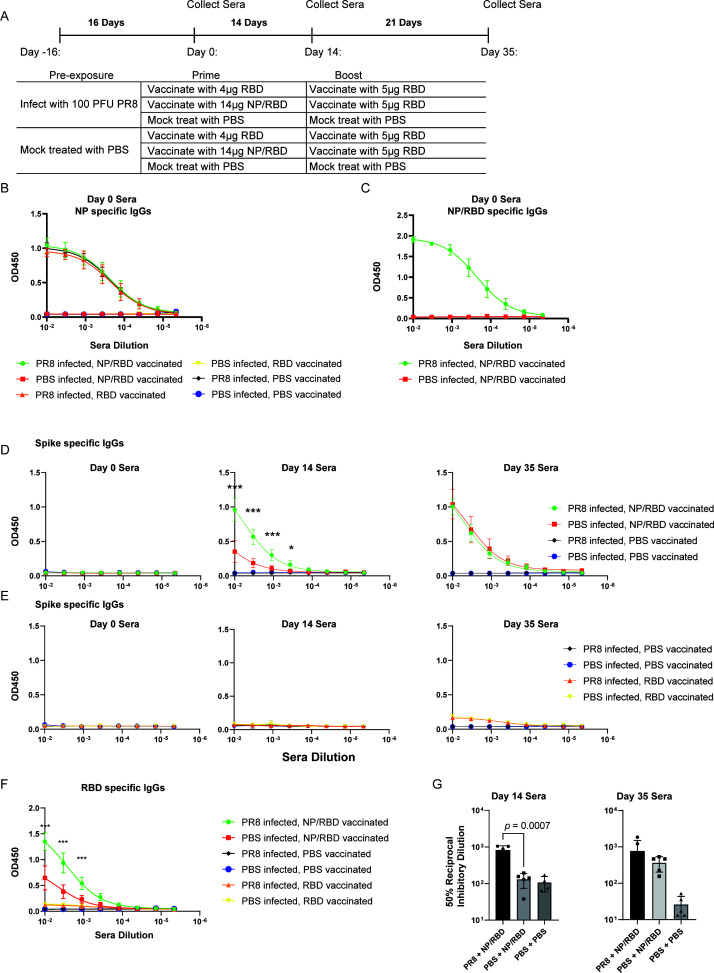
Antibody responses to vaccination with NP/RBD fusion protein in mice infected with PR8
To establish preexisting immunity to PR8 by viral infection, C57BL/6 mice were infected intranasally with 100 PFU of PR8 virus or phosphate-buffered saline (PBS). We knew from our previous work with influenza viruses that the mice would have robust anti-influenza virus adaptive immune responses between 14 and 21 days post-infection (DPI), so we chose this window of time for our initial vaccination studies. At 16 DPI, sera were collected, and the mice were vaccinated intramuscularly with equal molar amounts of either the NP/RBD fusion protein, RBD protein (amino acids 330–528), or PBS (Fig. 2A). As expected, our enzyme-linked immunosorbent assay (ELISA) analysis of sera from 16 DPI revealed strong NP antibody responses only from sera of animals infected with PR8 virus (Fig. 2B). These antibodies also recognized the NP/RBD fusion protein (Fig. 2C). To evaluate the RBD-specific antibody responses, we collected sera from mice at 14 days after vaccination and analyzed the sera by ELISA using plates coated with purified WA1 SARS-CoV-2 spike ectodomain protein. Vaccination with the NP/RBD fusion protein induced strong antibody responses to spike protein, while the purified RBD protein was poorly immunogenic (Fig. 2D and E). Antibody responses to SARS-CoV-2 spike protein were significantly increased in the mice previously infected with PR8 compared to mock-infected mice (Fig. 2D). This increase was observed when the ELISA was repeated with purified RBD protein as the ELISA antigen (Fig. 2F). We further tested the ability of the antibodies elicited by our NP/RBD vaccine to neutralize live WA1 SARS-CoV-2 virus. Antibodies from mice with preexisting immunity to influenza PR8 neutralized SARS-CoV-2 virus to a significantly greater extent than those from mice without preexisting influenza PR8 immunity (Fig. 2G).
Fig 2.
Antibody responses to vaccination with NP/RBD in mice previously exposed to influenza H1N1 PR8 virus infection. (A) Mouse vaccination schedule. (B) ELISA analysis of PR8 NP-specific IgG antibody levels in sera on Day 0 (n = 5/group). (C) ELISA analysis of NP-/RBD-specific IgG antibody levels in sera on Day 0 (n = 5/group). (D) ELISA analysis of RBD-specific IgG antibody responses induced by vaccination with NP/RBD or PBS in sera collected at Days 0, 14, and 35 after vaccination (ELISA performed with full-length spike ectodomain antigen) (n = 5/group). (E) ELISA analysis of RBD-specific IgG antibody responses induced by vaccination with RBD or PBS in sera collected at Days 0, 14, and 35 after vaccination (ELISA performed with full-length spike ectodomain antigen) (n = 5/group). (F) ELISA analysis of RBD-specific IgG antibody responses induced by vaccination with RBD or PBS in sera collected at Day 14 after vaccination (ELISA performed with RBD protein) (n = 5/group). (G) Plaque reduction assay analysis of neutralization potency on WA1 SARS-CoV-2 of IgG antibodies from sera collected at Days 14 and 35 after vaccination (***P ≤ 0.001; *P ≤ 0.05).
To evaluate whether spike-specific antibody responses could be further boosted by RBD subunit vaccination, we boosted the mice with our purified RBD protein (amino acids 330–528). We examined the spike-specific IgG responses at 3 weeks after boost (Day 35 after initial vaccination). Spike-specific antibodies were again high in the mice originally vaccinated with NP/RBD fusion protein (Fig. 2D and E). Surprisingly, antibodies to spike at this later time point were similar in the mice with or without exposure to influenza PR8 infection. This suggests that preexisting PR8 immunity accelerates RBD-specific antibody production.
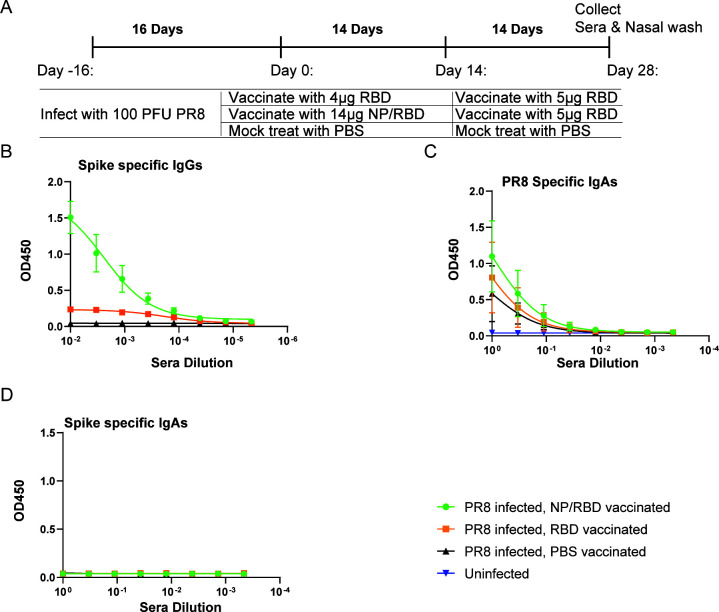
We were also interested in the mucosal IgA response elicited by our vaccine strategy in the nasal cavity, as the nasal cavity and upper airway are the initial sites of virus infection. Mice with preexisting immunity to influenza PR8 were vaccinated with NP/RBD fusion protein and boosted with RBD protein (Fig. 3A). We collected both sera and nasal wash samples 14 days after boost. ELISA results indicated that spike-specific antibodies in our sera samples remained high (Fig. 3B). In our nasal wash samples, we could detect influenza PR8-specific IgA antibodies but not spike-specific IgA antibodies (Fig. 3C and D). This was likely due to the different administration routes for PR8 virus and protein vaccination. Our protein vaccine was delivered intramuscularly while the mice had been infected with PR8 virus intranasally.
Fig 3.
Nasal wash IgA antibody responses. (A) Mouse vaccination regimen. (B) ELISA analysis of RBD-specific IgG antibody responses induced by vaccination in sera collected at Day 28 after vaccination (n = 4/group). (C) ELISA analysis of PR8-specific IgA antibody levels from nasal wash samples collected on Day 28 after vaccination (n = 4/group). (D) ELISA analysis of RBD-specific IgA antibody levels from nasal wash samples collected on Day 28 after vaccination (n = 4/group).
In summary, these results showed that vaccination with NP/RBD fusion protein triggers a neutralizing antibody response, which is significantly higher in animals with preexisting immunity to the influenza virus at an early time point, 14 days post vaccination.
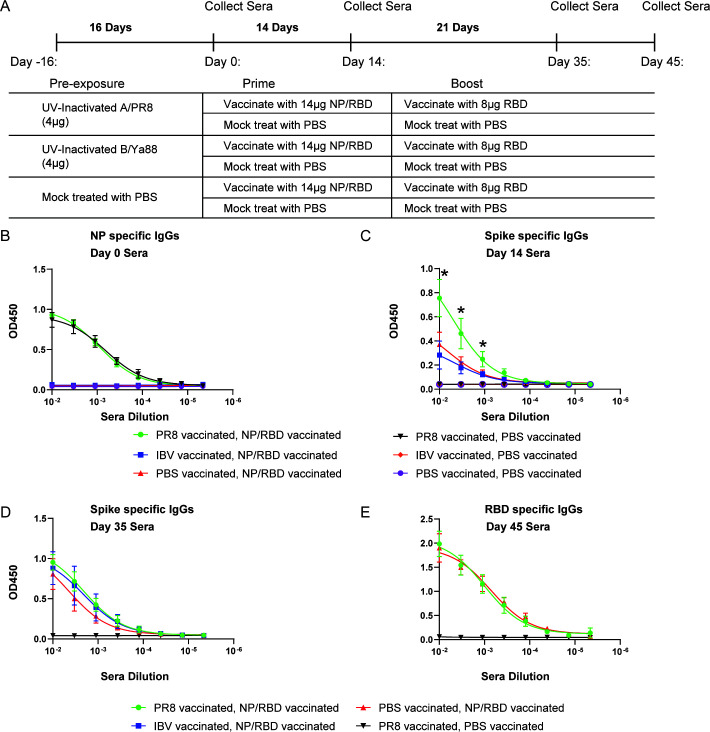
Antibody responses to NP/RBD vaccination in mice previously immunized with UV light-inactivated PR8
One alternative explanation for the increased antibody responses to RBD observed in Fig. 2 was that overall immune activation triggered by the virus infection drove the increase in antibody production. For example, increased cytokine production and priming of the innate immune system may have activated the adaptive immune system resulting in a faster antibody response following NP/RBD fusion protein vaccination. In this scenario, elevated antibody responses to RBD could be due to increased nonspecific immune responses.
To further test our interpretation that the enhanced antibody responses observed in Fig. 2 were a specific result from preexisting immunity to influenza PR8 virus, we established preexisting immunity in mice by immunizing with ultraviolet (UV)-inactivated influenza PR8 virus and used a group of mice treated with UV-inactivated influenza B/Victoria/2/1987 as a control (Fig. 4A). We included influenza B virus (IBV) because IBV NP and influenza A virus PR8 NP protein trigger distinct immune responses due to their low sequence similarities, about 30% amino acid identity.
Fig 4.
Antibody responses to vaccination with NP/RBD in mice previously vaccinated with UV-inactivated PR8. (A) Mouse vaccination regimen. (B) ELISA analysis of PR8 NP-specific IgG antibody levels in sera on Day 0. (C and D) ELISA analysis of RBD-specific IgG antibody levels in sera on Day 14 (C) or Day 35 (D) after vaccination (ELISA performed with full-length spike ectodomain antigen). (E) ELISA analysis of RBD-specific IgG antibody levels in sera on Day 45 after vaccination (ELISA performed with purified RBD protein) (n = 5/group for B–E) (*P ≤ 0.05).
At 16 days after vaccination with UV-inactivated virus, mice were vaccinated with either NP/RBD fusion protein or PBS. Sera were collected at Day 0 and tested for PR8 NP-specific antibodies. ELISA results indicated PR8 NP-specific antibodies were detected in sera from mice treated with UV-inactivated PR8 virus but not from mice treated with UV-inactivated IBV (Fig. 4B). This confirmed the divergence between these two NP proteins. At 14 days after vaccination with NP/RBD fusion protein, we collected sera to evaluate the spike-specific antibody responses. Similar to our previous result using live virus infection, we observed that antibody responses to spike were significantly increased in mice that had preexisting immunity to PR8 at Day 14 but not in mice that had preexisting immunity to IBV or PBS (Fig. 4C). This demonstrates that the observed increase in spike-specific antibodies is the result of specific preexisting immunity to PR8.
To evaluate whether the RBD-specific antibodies could be further enhanced by RBD vaccination, we further vaccinated the mice with our purified RBD protein (amino acids 330–528). We assessed spike and RBD-specific antibodies in sera samples collected at both Days 35 and 45 after vaccination. ELISA results indicated abundant RBD-specific antibody responses but without significant differences between mice with or without previous exposure to influenza PR8 virus (Fig. 4D and E). These results suggest that vaccination with NP/RBD fusion protein can harness preexisting immunity established by a specific previous exposure to influenza virus to accelerate antibody responses to SARS-CoV-2 spike protein after a single vaccination dose.
Splenocyte stimulations from mice vaccinated with NP/RBD fusion protein
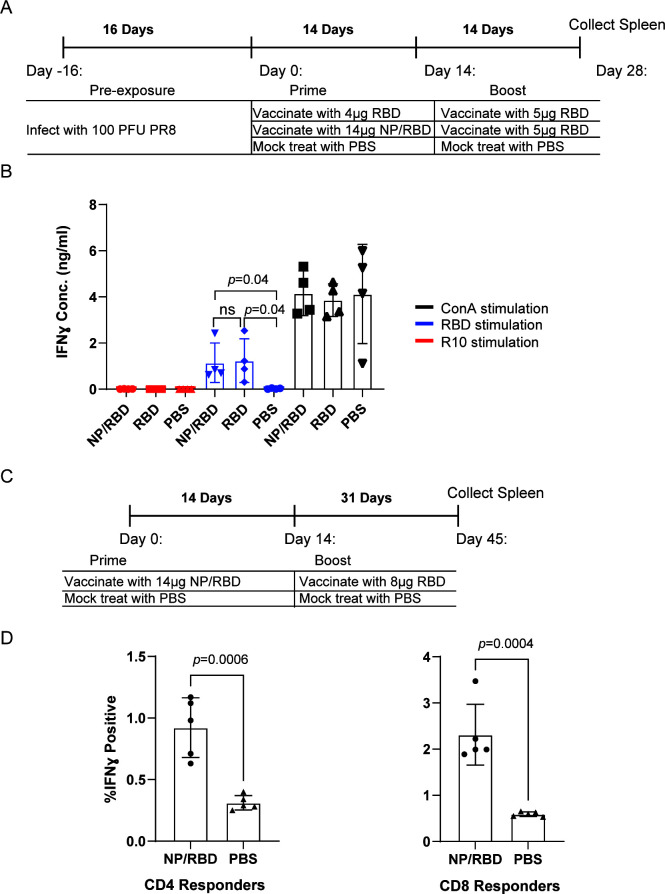
T-cell responses are an important protective adaptive immune response in addition to antibodies. We therefore wished to check if our vaccine regimen with NP/RBD fusion protein produced RBD-specific T-cell responses. Mice that were vaccinated with either NP/RBD or RBD protein followed by boosting with RBD protein were sacrificed 2 weeks after boost, and their spleens were collected for splenocyte stimulation assays (Fig. 5A). RBD protein was used to stimulate splenocytes, and interferon gamma (IFNγ) production was measured by ELISA as an output for RBD-specific T-cell activation. Concanavalin A was used as a positive control to nonspecifically activate T cells. Both vaccine groups showed comparable splenocyte activation following the stimulation with RBD protein over unvaccinated mice, and there was no difference in T-cell activation between the NP/RBD and RBD vaccine groups (Fig. 5B). To confirm the observation of RBD-specific T-cell responses, we performed a separate experiment where mice vaccinated with NP/RBD fusion protein followed by boosting with RBD were sacrificed 31 days after boost (Fig. 5C). RBD protein was used to stimulate splenocytes, and IFNγ production was measured by intracellular cytokine staining. Flow cytometric analysis of stained lymphocytes showed significantly increased RBD-specific CD4- and CD8-positive cells in mice vaccinated with NP/RBD fusion protein compared to unvaccinated mice (Fig. 5D; Fig. S2). Taken together, these results demonstrate that a vaccine strategy that utilizes NP/RBD is capable of activating RBD-specific T-cell responses, in addition to RBD-specific antibody responses.
Fig 5.
T-cell responses to vaccination with NP/RBD followed by boosting with RBD. (A) Mouse vaccination regimen for data in B. (B) Evaluation of activated RBD-specific splenocyte responses using IFNγ ELISA with the supernatants from splenocytes taken from Day 28, stimulated with RBD, ConA as a positive control, or R10 media as a negative control. (C) Mouse vaccination regimen for data in D. (D) Evaluation of activated RBD-specific T-cell responses. Intracellular cytokine staining for IFNγ after RBD stimulation of CD4- and CD8-positive lymphocytes from mice vaccinated with either NP/RBD and boosted with RBD or mice vaccinated with PBS (mock).
NP/RBD vaccination in mice preexposed to inactivated PR8 protects mice against SARS-CoV-2-induced morbidity and mortality
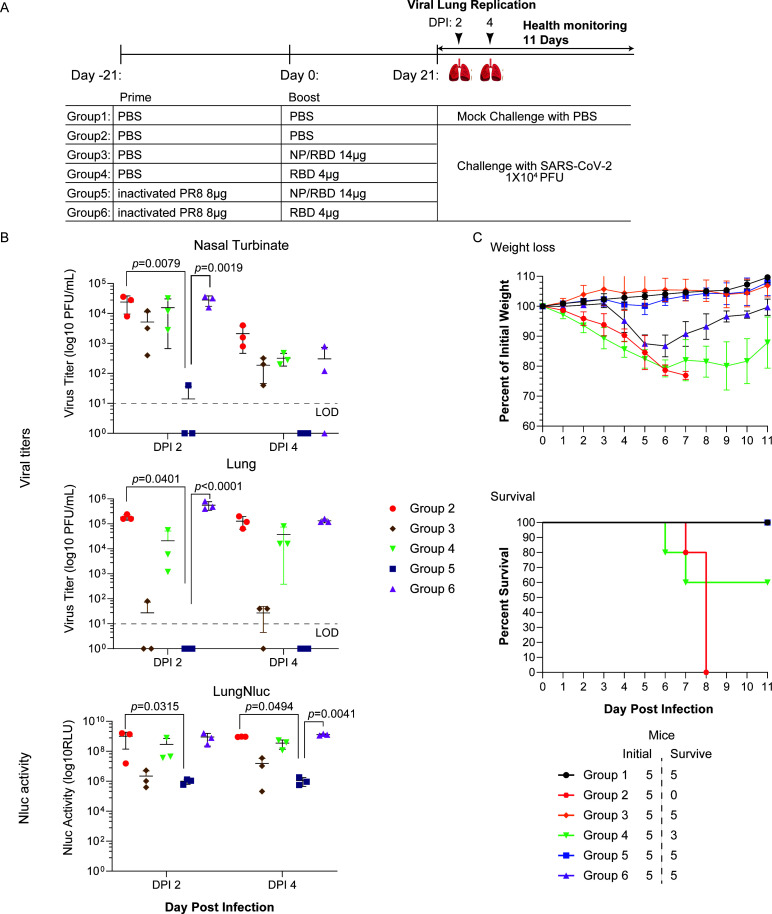
To further test whether our vaccine regiment could induce protective immunity against live virus infection, we performed a challenge study using recombinant SARS-CoV-2 expressing mCherry and nanoluciferase (rSARS-CoV-2/mCherry-Nluc) at a dose of 104 PFU/mouse (39). fK18 hACE2 transgenic mice were first primed with either PBS or inactivated PR8 virus and then subsequently vaccinated with either NP/RBD or RBD and challenged with rSARS-CoV-2 (Fig. 6A). At 2 and 4 DPI, three mice from each group were sacrificed to determine virus replication. Noticeably, we observed the lowest reading for the in vivo bioluminescence imaging, pulmonary Nluc bioluminescence and mCherry fluorescence signals, and virus titers in the cohort that was preexposed to PR8 and vaccinated with the NP/RBD protein (Fig. 6B; Fig. S3A and S3B; Table S1). This suggested that this vaccine regimen accelerated rSARS-CoV-2 virus clearance in both upper (nasal turbinate) and lower (lungs) respiratory tracks. Moreover, we monitored the health of the challenged mice up to 11 DPI. We observed that only the two cohorts vaccinated with NP/RBD were completely protected against weight loss and death from lethal challenge with rSARS-CoV-2 (Fig. 6C). Together, our results suggest that NP/RBD vaccination in mice with preexposure to PR8 is protective against SARS-CoV-2 replication and subsequent morbidity and mortality.
Fig 6.
SARS-CoV-2 challenge of NP/RBD-vaccinated mice. (A) The vaccination and challenge regimen for female 4- to 6-week-old K18 hACE2 transgenic mice. (B) Quantification of virus concentrations in the lungs and nasal turbinate by plaque assay and Nluc activity in the lungs. The limit of detection (LOD) is 10 PFU/mL. (C) Body weight changes and percent mortality were monitored daily up to 11 DPI (n = 5/group). Data are presented as mean ± SEM.
Comparison of sera antibody responses in mice vaccinated with NP/RBD or RBD proteins
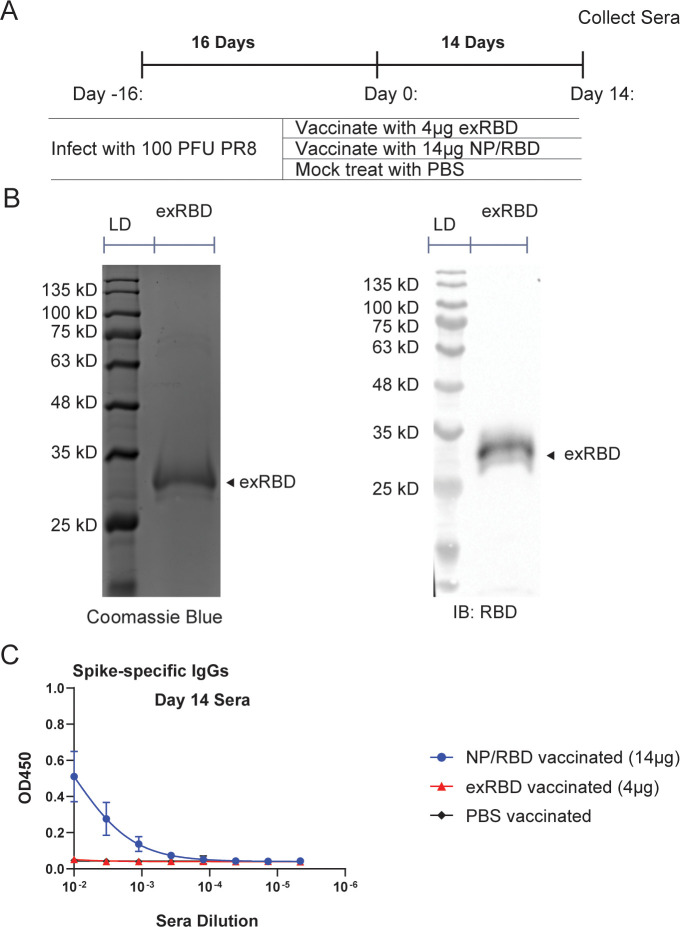
The poor immunogenicity of our RBD protein was surprising since many reports have shown RBD protein is a suitable vaccine candidate for SARS-CoV-2 (26, 40, 41). We speculated that this may have been due to its poor glycosylation because our design covers amino acids 330–528, resulting in the inclusion of a glycosylation site at amino acid position 331 very close to the N-terminus of our protein (42, 43). Since it is known that glycosylation can affect antibody responses to antigens (44), we therefore wished to use an extended version of RBD (exRBD) containing amino acids 319–528 for mouse vaccinations (Fig. 7A). We purified the exRBD protein and verified the protein purification by Western blot and Coomassie stain (Fig. 7B). At 14 days after vaccination with exRBD or our original NP/RBD protein, sera were collected, and IgG antibody responses to spike were determined by ELISA. The NP/RBD fusion protein again produced greatly increased antibody responses compared to exRBD, while the purified exRBD protein was again not immunogenic (Fig. 7C). In summary, vaccination with the NP/RBD fusion protein shows superior immunogenicity compared to vaccination with RBD alone, which is likely due to the potential stabilization role of NP in maintaining RBD folding or oligomerization of NP/RBD fusion protein (Fig. S1).
Fig 7.
Comparison of RBD-specific antibody responses from sera of mice vaccinated with either NP/RBD or exRBD (amino acids 319–528). (A) Mouse vaccination regimen. (B) Coomassie blue stain (left) and Western blot (right) of purified exRBD 6xHis (amino acids 330–528) (LD = protein ladder). (C) ELISA analysis of RBD-specific IgG antibody levels in sera on Day 14 after vaccination (n = 5/group).
CD4-positive T cells in mice with preexisting immunity to PR8, but not preexisting antibody responses, are necessary for accelerated RBD antibody responses after NP/RBD vaccination
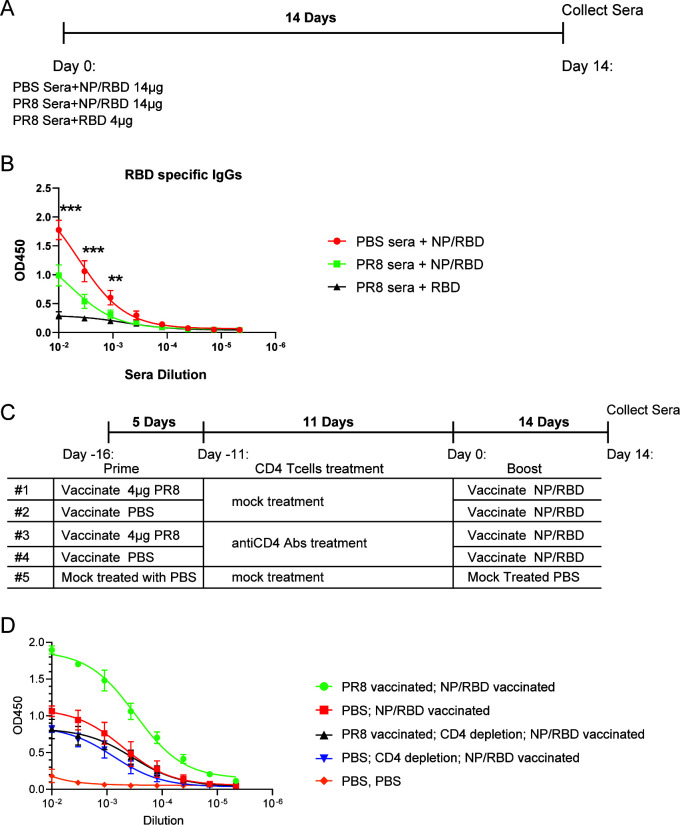
Our results indicated that preexisting adaptive immunity to influenza PR8 virus accelerated the production of spike-specific IgG antibodies. There are two main broad components of the adaptive immune response that could account for these results: T-cell responses and antibody responses. To test the potential role of antibody responses to PR8 involved in the increase in RBD-specific antibodies, sera from the mice in Fig. 2A collected on Day 16 after infection were pooled together and incubated with NP/RBD fusion protein for 1 hour at 4°C. This allowed for the binding of anti-NP antibodies in the sera to the NP/RBD fusion protein. Sera from mice treated with PBS were used as a control. After incubating the sera with NP/RBD fusion protein, the mixture was combined with adjuvant and used for vaccinations (Fig. 8A). Fourteen days after vaccination, sera were collected from the mice, and RBD-specific IgG antibodies were determined by ELISA. Interestingly, mice vaccinated with NP/RBD pretreated with PR8 sera had significantly lower antibody responses compared to mice vaccinated with NP/RBD pretreated with PBS sera (Fig. 8B, P = 0.0001 for first dilution). This result indicated that the antibody response to influenza is not responsible for the increased RBD responses observed in Fig. 2 and 4 following NP/RBD vaccination.
Fig 8.
Comparison of RBD-specific antibody responses in mice vaccinated with NP/RBD 6xHis protein treated with PR8 sera or PBS sera. (A) Mouse vaccine regimen. (B) ELISA analysis of RBD-specific IgG antibody levels in sera from A on Day 14 after vaccination (n = 5/group) (***P ≤ 0.001; **P ≤ 0.01). (C) Mouse vaccine regimen with or without CD4 T-cell depletion. (D) ELISA analysis of RBD-specific IgG antibody levels in sera from C on Day 14 after vaccination (n = 5/group).
To test the hypothesis that CD4-positive T cells in mice with preexisting immunity to PR8 were responsible for the increased RBD responses observed after NP/RBD vaccination, we performed vaccinations using the same regimen as described in Fig. 4A and included two vaccine groups with depleted CD4 T cells using anti-mouse CD4 monoclonal antibodies (clones GK1.5, BioXCell) after their initial exposure to PR8 viruses (Fig. 8C). Their CD4 T-cell populations were reconstituted as shown in Fig. S6. In the animals that received the CD4 depletion treatment, the results showed similar RBD-specific antibody responses in the two groups, whether or not they had prior exposure to the PR8 virus (Fig. 8D). However, for mice without CD4 T-cell depletion treatment, we observed the similar enhanced RBD antibody responses in the animals with prior exposure to PR8 virus compared to the ones without PR8 exposure after the NP/RPD vaccination (Fig. 8D). These results suggest that the enhancement of RBD-specific antibodies is dependent on the presence of PR8-specific CD4 T cells.
DISCUSSION
The results obtained in this study demonstrate that antibody responses to a novel viral pathogen can be accelerated by drawing on previous immunity to an unrelated pathogen. We devised a vaccine strategy to utilize preexisting immunity to influenza virus with the goal of increasing antibody responses to SARS-CoV-2. We purified a fusion protein of influenza NP protein and SARS-CoV-2 spike RBD protein as a vaccine candidate against SARS-CoV-2. Vaccination with the NP/RBD fusion protein produced high levels of spike-specific antibodies after a single dose, which were significantly higher in mice with preexisting immunity to influenza virus as early as 2 weeks after vaccination. Vaccination with NP/RBD fusion protein also produced significantly higher antibody responses compared to equal-molar vaccinations with RBD protein alone, which was not greatly immunogenic under our vaccination conditions. Furthermore, antibody responses to NP/RBD fusion protein in mice with preexisting immunity to influenza virus exhibited greater neutralization of SARS-CoV-2 virus than antibodies produced in mice without preexisting immunity. Additionally, NP/RBD vaccination followed by boosting with RBD protein was shown to produce RBD-specific splenocyte activation and durable RBD-specific T-cell responses. Mice vaccinated with a single dose of NP/RBD were protected against SARS-CoV-2-induced morbidity, mortality, and viral replication, while vaccination with RBD was only partially protective against mortality and did not protect against morbidity. Additionally, when we quantified virus replication in the nasal turbinate and lungs of infected mice, the mice that were preexposed to influenza virus and then vaccinated with NP/RBD had the lowest viral titers among all tested groups. Mechanistically, our results indicated that CD4-positive T cells in mice with preexisting immunity to PR8 influenza virus were responsible for the accelerated RBD antibody response after NP/RBD vaccination. Collectively, these results show the benefits of an NP/RBD vaccine candidate against SARS-CoV-2 and of utilizing preexisting immunity to boost antibody responses against a novel pathogen.
While we had hypothesized that preexisting helper T-cell immunity to NP could boost antibody responses to RBD after NP/RBD fusion protein vaccination, preexisting B-cell and antibody responses to NP could have been expected to decrease antibody responses to RBD after NP/RBD vaccination (45–48). We speculated that NP-specific B cells expanded during PR8 immunization could outcompete RBD-specific B cells for access to NP/RBD antigen and for space in lymphatic tissue. Another possibility suggested that antibodies binding to NP epitopes might sterically hinder the binding of B-cell receptors that recognize novel epitopes (48). Also, it is possible that NP-specific antibodies bound to NP/RBD could reduce the movement of the NP/RBD protein to primary lymphoid tissues and germinal centers (49). Our results in Fig. 8B showed lower RBD-specific antibodies after vaccination with NP/RBD fusion protein mixed with sera from influenza virus-infected mice. This observation suggests that NP-specific antibodies negatively impact RBD antibody responses after NP/RBD vaccination and favored our hypothesis that NP-specific helper T cells were responsible for the increased RBD antibody responses after NP/RBD vaccination.
To strengthen this conclusion, we further directly assessed the role of preexisting NP-specific CD4 T cells in accelerating RBD antibody responses by temporarily depleting CD4 cells after the initial exposure to influenza virus. We showed that the transient depletion of CD4-positive T cells in mice with preexisting immunity to PR8 negated the previously observed accelerated antibody response against RBD following NP/RBD vaccination. The results indicated that the enhancement of RBD-specific antibody responses in mice with preexisting immunity to PR8 is likely due to the presence of expanded NP-specific helper T cells, and we surmise that the presence of this expanded cell population is enough to overcome any potential negative impact of NP-specific antibodies or B cells, under our vaccine conditions. The vaccination regimen may play a role in this observation. Specifically, the outcome might be influenced by the timing between the primary exposure to PR8 virus and the subsequent NP/RBD vaccination. Extending the time interval between primary vaccination and booster generally has a positive effect on antibody responses across different vaccine platforms, at least if it is within a 6-month time post-primary vaccination (50, 51). However, adaptive immune responses decline over time, noticeably 6 months after vaccination in SARS-CoV-2 and influenza vaccine studies (31, 52). Vaccination with NP/RBD at a later time point following exposure to influenza antigen may or may not result in the increases we observed, especially as the effector NP-specific T-cell populations decline. Nevertheless, the results we obtained in this study favor our conclusion that at least under some circumstances, preexisting immunity can be used to optimize vaccine immune responses toward a divergent virus. The role of NP antibody and B-cell responses, preexisting NP-specific T cells, and the timing of vaccinations will be the focus of future studies designed to better understand how preexisting immunity interacts with the chimeric vaccine protein.
We were surprised at the poor immunogenicity of the RBD protein compared to NP/RBD fusion protein, since RBD protein has been shown to be a suitable SARS-CoV-2 vaccine candidate (19, 21, 23, 26). These observed differences in RBD immunogenicity might be explained by differences in vaccine doses, mouse strains, and amino acid composition of the RBD antigen used in this study. The ability of influenza NP to form oligomers may contribute to the increased immunogenicity of the NP/RBD fusion protein over monomer RBD, and many vaccine platforms with RBD have made use of dimerized RBD or incorporated RBD into nanoparticle structures (20, 22, 23, 26). The potential oligomerization of NP/RBD fusion protein could enhance its stability and allow for crosslinking of B-cell receptors upon antigen recognition for greater B-cell stimulation (22, 53). This enhanced immunogenicity of NP/RBD from its structure as well as its engagement with immune responses to influenza makes it an ideal vaccine candidate.
The vaccine strategy outlined in this report can, in theory, be incorporated or combined with other vaccine platforms, such as mRNA or DNA vaccines. It is worth exploring vaccine strategies that draw on immunity to pathogens other than influenza viruses. In this regard, virus-like particle vaccines based on fusion proteins with the hepatitis B virus surface antigen (HBsAg) have shown some similarity to the responses seen with NP/RBD fusion protein. In humans and in animals, preexisting immunity to HBsAg can increase antibody responses against Plasmodium falciparum antigens (54, 55), although the mechanism behind this increase has not been shown. There are broad applications for the strategy of targeting preexisting immunity to increase antibody responses to novel pathogens, such as new SARS-CoV-2 variants of concern, novel coronaviruses, or emerging zoonotic viruses. In summary, our results demonstrate the beneficial effects of using an NP/RBD vaccine against SARS-CoV-2. This provides a paradigm for vaccine design to generate accelerated immune responses against novel viral pathogens with the help of preexisting immunity against a separate viral pathogen.
MATERIALS AND METHODS
Cell lines and mice
XL1-blue Escherichia coli were used to produce the pFastBac plasmids for baculovirus rescue. SF9 insect cells grown in ESF 921 media (Expression Systems) were used for baculovirus rescue and expansion and for the production of vaccine proteins. 293T cells were used for the production of RBD (amino acids 319–528) protein. Vero cells were used for SARS-CoV-2 neutralization assays. Both cells were cultured with Dulbecco's modified Eagle's medium (DMEM) (Corning) containing 10% fetal bovine serum (FBS) (Thermo Fisher). The animal studies in this paper made use of either 6-week-old female C57BL/6 mice purchased from Jackson Labs or 4- to 6-week-old female K18 hACE2 transgenic mice (B6.Cg-Tg(K18-ACE2)2Prlmn/J) from the Jackson Laboratory. C57BL/6 mice were maintained in the specific pathogen-free animal facility at the University of California, Riverside, and the K18 hACE2 mice were maintained at Texas Biomed.
Virus strains
Mouse infections were performed with the A/Puerto Rico/8/34 (H1N1) influenza virus strain. Mouse vaccinations with UV-inactivated virus were performed with A/Puerto Rico/8/34 (H1N1) and B/Victoria/2/1987 influenza virus strains. SARS-CoV-2 neutralization assays were performed with USA-WA1/2020 SARS-CoV-2 strain. The rSARS-CoV-2/mCherry-Nluc was previously described (39). Viral stocks were generated in Vero E6 cells, aliquoted, titrated, sequenced and stored at −80°C as previously described (39).
Plasmids
The genes for the extracellular domain of WA1 spike protein (amino acids 22–1208), WA1 spike RBD domain (amino acids 330–528), and NP/RBD were cloned into the pFastBac plasmid for baculovirus rescue with an N-terminal secretion signal sequence derived from the gene for influenza A/WSN/33 H1N1 (WSN) hemagglutinin and a C-terminal 6xHis tag for protein purification. pFastBac plasmid was digested with HindIII and BamHI restriction enzymes, and genes of interest were inserted into the plasmid by InFusion ligation. Plasmid sequences were verified by Sanger Sequencing, and purified plasmids were used for baculovirus rescue.
The gene for RBD (amino acids 319–528) was cloned into the pCAGGS plasmid with an N-terminal secretion signal derived from the gene for WSN influenza hemagglutinin and a C-terminal 6xHis tag for protein purification. pCAGGS plasmid was digested with EcoRI and XhoI restriction enzymes, and the RBD gene was inserted into the plasmid by InFusion ligation. Plasmid sequences were verified by Sanger Sequencing, and purified plasmids were used for protein expression in 293T cells.
Baculovirus generation
Baculovirus rescues for protein expression were performed using the Bac-to-Bac Baculovirus Expression System according to the manufacturer’s instructions (Invitrogen). Rescued virus was amplified twice in SF9 cells to produce P2 virus. Western Blots were performed with the cells used to produce the P2 virus to verify the expression of the protein of interest. Verified P2 stocks were used to infect SF9 cells for protein purification.
Protein purification
WA1 spike, spike RBD (amino acids 330–528), and NP/RBD proteins were purified with the baculovirus expression system. Two hundred milliliters of SF9 cells at a density of 1 × 106 cells/mL were infected with 20 mL of P2 baculovirus containing the gene of interest. Forty-eight hours after infection, the cells were collected and spun down at 500 g for 5 minutes. The supernatants were collected and passed through a 0.2-micron filter, and phenylmethylsulfonyl fluoride (PMSF) protease inhibitor was added to the filtrate to a final concentration of 1 mM. The filtrate was then passed through a 5-mL HisTrapFF Ni column (Cytiva) at a flow rate of 5 mL/minute. The column was washed with 50 mL of wash buffer containing 20-mM imidazole, 300-mM NaCl, and 50-mM monobasic sodium phosphate. Protein was eluted from the column with 20 mL of elution buffer containing 250-mM imidazole, 300-mM NaCl, and 50-mM monobasic sodium phosphate. Eluted protein was diafiltrated and concentrated with a 15-mL 10-kD cutoff column (Amicon Ultracel 10K centrifugal filters), and the column was washed twice with PBS. Protein concentration was determined by Bradford Assay.
Spike RBD (amino acids 319–528) protein and NP/RBD protein were purified by transfection of 293T cells with the RBD-pCAGGS plasmid or NP/RBD-pCAGGS plasmid. Five T175 flasks (Genesee Scientific) of fully confluent 293T cells were polyethylenimine (PEI) transfected with 30 µg of plasmid in Opti/MEM media (Thermo Fisher) at a ratio of 3-µL PEI:1-µg DNA. Six hours after the addition of DNA/PEI to the cells, the medium was changed to 20-mL DMEM media per flask containing 10% FBS, and cells were left at 37°C for 48 hours. After 48 hours, the supernatant was collected, and protein was purified with a 5-mL HisTrap column (Cytiva). Eluted protein was diafiltrated with a 10-kD cutoff column (Amicon Ultracel 10K centrifugal filters) and washed twice with PBS. Protein concentration was determined by Bradford Assay, and purify was determined by Coomassie blue stain. Protein purified from the HisTrap column was used for all vaccination studies.
The eluted protein was further purified by size exclusion chromatography with a Superdex 200 16/600 column (GE Healthcare) preequilibrated with buffer containing 25-mM Tris, pH 7.5, 500-mM NaCl, 5-mM DTT, and 5% glycerol through an AKTAgo system. Protein samples were collected at 1 mL/fraction, and fractions from the same chromatogram peak were pooled together. The fraction pool corresponding to the RBD-containing protein was collected and verified by Western blot and Coomassie Blue stain. Protein concentration was determined by Bradford Assay.
Western blots and Coomassie blue stains
Western blots for RBD-containing proteins were blocked with 2% bovine serum albumin (BSA) in PBS containing 0.1% Tween20 (PBST) and then blotted with anti-SARS-CoV-2 spike RBD Monoclonal Antibody (R&D Systems, Catalog # MAB10540) diluted 1:500 in 2% BSA/PBST for 1 hour. Blots were then incubated with goat α-mouse IgG-HRP secondary antibody (Prometheus) diluted 1:10,000 in blocking buffer for 1 hour. Blots were visualized with the Bio-Rad ChemiDoc Touch Imaging System. Coomassie gels were stained as previously described (56).
AlphaFold prediction
Predicted protein structures for the RBD (amino acids 330–528) and NP/RBD were determined with the AlphaFold Colab based on AlphaFold v2.1.0 using the AlphaFold model parameters (38).
Mouse vaccinations and influenza infections
C57BL/6 mice anesthetized with isoflurane were infected intranasally with 100 PFU of PR8 virus (A/Puerto Rico/8/34 strain) in 50-µL PBS containing 0.4% BSA and 1% Pen-Strep antibiotic to establish preexisting immunity to PR8. Purified virion of PR8 and IBV (B/Victoria/2/1987 strain) diluted in PBS was UV-inactivated on ice for 30 minutes and added to an equal volume of AddaVax adjuvant (InvivoGen) for vaccinations. NP/RBD protein and RBD (amino acids 330–528) purified from SF9 cells were combined with equal volume AddaVax adjuvant before vaccination. For the vaccinations in Fig. 8 done with sera-treated NP/RBD, 117 µL of NP/RBD protein (140 µg) or RBD protein was combined with 133 µL of sera and incubated at 4°C for 1 hour. An equal volume of AddaVax adjuvant was then added to the sera/protein mixture per the manufacturer’s instructions, and the resulting solution was used for vaccinations. For all vaccinations, mice were injected intramuscularly with 50 µL of vaccine per injection in the thigh muscle of both back legs. Equal molar amounts of NP/RBD and RBD were used in all vaccinations.
Mouse SARS-CoV-2 challenge
For viral challenge studies, groups of vaccinated K18 hACE2 transgenic mice were anesthetized with gaseous isoflurane and inoculated intranasally with 104 PFU/mouse of rSARS-CoV-2/mCherry-Nluc (39). A separate group of K18 hACE2 transgenic mice were also mock-infected with PBS and served as a negative control. For body weight and survival studies, K18 hACE2 transgenic mice (n = 5) were anesthetized with gaseous isoflurane and intranasally infected with 104 PFU/mouse of rSARS-CoV-2/mCherry-Nluc and monitored daily for body weight loss and survival to access morbidity and mortality, respectively, for 11 days. Mice that were below 75% of their initial body weight were considered to have reached their experimental endpoint and were humanly euthanized. In vivo bioluminescence imaging of live mice (n = 3) was conducted with an Ami HT in vivo imaging system (IVIS; Spectral Instruments) at 2 and 4 DPI (39). Mice were anesthetized with isoflurane and retro-orbitally injected with 100 µL of Nano-Glo luciferase substrate diluted by 1:10 in PBS (39). Mice were immediately placed in an isolation chamber and imaged using the Ami HT IVIS. Radiance within the region of interest of each mouse was analyzed using the Aura software (Spectral Instruments), and total flux values (protons/s) were normalized to the background signal of mock-infected control. To access bioluminescence (Nluc) and fluorescence (mCherry) expression in the lungs and to determine viral titers, mice from the bioluminescence imaging studies (n = 3) were humanely euthanized at 2 and 4 DPI after in vivo imaging (39). Lungs were surgically excised and washed in PBS, and Nluc, mCherry, and brightfield images were obtained using an Ami HT IVIS (Nluc and mCherry) and an iPhone 6s (Apple, brightfield images). Tissue homogenates were centrifuged at 12,000 × g at 4°C for 5 minutes to pellet cell debris, and supernatants were collected. Viral titers were determined using standard plaque assay and immunostaining using a SARS-CoV nucleocapsid (N) protein cross-reactive antibody (1C7C7). Nluc activity in the lung tissue homogenates was also determined using Nano-Glo luciferase substrate kit and a GloMax Navigator microplate luminometer.
Sera and nasal wash collections
Retroorbital blood was collected from mice with a Pasteur pipet. The blood was spun down at 10,000 g for 10 minutes to separate the sera. Sera was collected and stored at −80°C. Nasal washes were performed after mouse euthanasia with CO2. A pipet tip was inserted into the trachea, and 1 mL of PBS was back-washed through the nose from the trachea. The collected nasal wash was stored at −80°C.
Enzyme-linked immunosorbent assays
ELISAs to determine IgG antibody responses to PR8 NP protein, SARS-CoV-2 spike protein, and SARS-CoV-2 spike RBD protein were performed as follows. MaxiSorp 96-well ELISA plates (Thermo Fisher) were coated with 50 µL of purified target antigen at a concentration of 2.5 µg/mL. Antigen was allowed to bind to the plate overnight at 4°C, and then the plate was blocked with a blocking buffer (1% milk in PBST). Sera was diluted 1:100 in blocking buffer and then serially diluted threefold. Plates were incubated with sera for 2 hours, after which the plates were washed three times with a blocking buffer. Plates were incubated with goat α-mouse IgG-HRP secondary antibody (Millipore Sigma AP503P) diluted 1:3,000 in a blocking buffer and incubated for 1 hour. Plates were washed three times with blocking buffer and then incubated with 100 µL of SigmaFast OPD substrate (Cat # P9187) for 30 minutes. The substrate reaction was stopped with 25 µL of 3-M HCl, and absorbance was measured at 450 nm with a Luminometer plate reader (Promega). ELISAs to determine IgA antibody responses were performed in a similar manner. The starting dilution of nasal washes was undiluted sample, followed by serial threefold dilutions in blocking buffer. Plates were incubated with nasal wash for 2 hours, washed with a blocking buffer three times, and then incubated with goat anti-mouse IgA-HRP antibody (Southern Biotech) diluted 1:2,000 in blocking buffer for 1 hour. Substrate addition and absorbance measurements were performed as with IgG responses.
SARS-CoV-2 neutralization assays
Sera used for neutralization assays were treated with the receptor-destroying enzyme (RDE) overnight at 37°C, and then RDE was inactivated at 56°C for 30 minutes. Sera were serially diluted 1:3 in a 96-well plate with PBS, and 100 µL of sera was combined with 50 PFU of WA1 SARS-CoV-2 virus (57) in 100 µL diluted in PBS. Sera were allowed to bind to virus for 1 hour at room temperature, and then the virus + sera solutions were used to infect Vero cells in 12-well plates for 1 hour at 37°C with shaking every 5 minutes. The medium was then changed to plaquing media (DMEM + 1% Avicell + 2% FBS + 1% P/S) (58). Cells were incubated for 72 hours at 37°C. The cells were then fixed with 3.7% formaldehyde for 1 hour, then washed with water, and stained with crystal violet for 45 minutes. The plaques for each sera dilution were calculated and compared to no-sera controls to determine plaque reduction; 50% sera reciprocal inhibitory dilutions were calculated as previously described (59).
Splenocyte stimulations
Mice were euthanized with carbon dioxide on the day of splenocyte collection. Spleens were mashed through a 40-micron cell strainer and washed with 10-mL R10 media (RPMI media containing 1% glutamine, 1% Pen/Strep, 1% HEPES, and 10% FBS). Splenocytes were spun down at 1,200 rpm for 10 minutes and then resuspended in 3 mL of ammonium–chloride–potassium (ACK) red blood cell lysis buffer (150-mM NH4Cl, 10-mM KHCO3, 0.1-mM EDTA, and pH 7.2) for 5 minutes, after which 10 mL of R10 media was added to the cells, and the cells were spun down again at 1,200 rpm for 10 minutes. Cells were resuspended in 4 mL of R10 media and counted with a hemocytometer; 3 × 106 cells/sample were added to each well of a 96-well plate in 50 µL of R10 media. An additional 50 µL of R10 media, R10 media containing RBD at 30 µg/mL, or R10 media containing 2 µg/mL Concanavalin A (ConA) (Sigma Aldrich) was added to the cells. Cells were stimulated for 48 hours for IFNγ ELISAs or overnight for intracellular cytokine staining.
Intracellular cytokine staining and flow cytometric analysis
Stimulated splenocytes were collected and washed twice with 200-µL FACS buffer (PBS, 2-mM EDTA, 3% FBS). Cells were blocked with Fc block (BD 553142) for 5 minutes and then stained with CD4, CD8, and CD3 antibodies for 30 minutes at room temperature: anti-mouse CD8a FITC (Invitrogen 11-0081-81), anti-mouse CD4 APC-eFlour780 (Invitrogen 47-0041-82), and anti-mouse PerCP-Cyanine 5.5 (Invitrogen 45-0031-82). Cells were then washed twice with 200-µL FACS buffer. Cells were fixed and permeabilized with eBioscience fixation/permeabilization solution (00-5123) and then stained with anti-mouse IFNγ PE (Invitrogen 12-7311-81) for 30 minutes at room temperature. Cells were washed twice with 200-µL FACS buffer and analyzed by flow cytometry with a BD FACSCantoII system recording 60,000 total events for each sample. Gating and sample analysis were performed with FlowJo software v10.8.
CD4-positive T-cell depletion
To deplete CD4-positive T cells, anti-mouse CD4 monoclonal antibodies (clones GK1.5, BioXCell) were injected intraperitoneally at a dose of 125 µg/mouse on Day 5 following the initial vaccination with inactivated PR8 virus (Fig. 8C) (60). Control animals received an equivalent amount of BSA (Fig. 8C). To assess the different T-cell populations, spleens were isolated from mice at the indicated time points, and single-cell suspensions were obtained by mechanical homogenization. Freshly isolated cells were stained with the antibodies purchased from eBioscience (San Diego, CA, USA) or BD Biosciences (San Jose, CA, USA) and were run on a FACSCalibur (BD, San Jose, CA, USA).
Statistical analysis
Unpaired t-tests for significance for the T-cell stimulations, multiple unpaired t-tests for significance for the ELISAs, and two-way analysis of variance with Tukey’s multiple comparison test for significance for the viral titers in respiratory tissues were performed with GraphPad Prism 9.2.0 software.
ACKNOWLEDGMENTS
We would like to thank Tran Phan for her assistance setting up the BSL3 laboratory at UCR. We are also grateful for Dr. Chengjin Ye, who provided detailed technical support to generate SARS-CoV-2 virus stocks.
This work was partially supported by NIAID (1R21AI147057 and R01AI153419) to R.H. and NIH (NS072298 and RAI124682) to E.H.W. Research on SARS-CoV-2 in LM-S laboratory was partially supported by grants W81XWH2110103 and W81XWH2110095 from the Department of Defense (DoD) Peer Reviewed Medical Research Program (PRMRP); 1R43AI165089-01, 1R01AI161363-01, and 1R01AI161175-01A1 from the National Institute of Health (NIH); the Center for Research on Influenza Pathogenesis (CRIP), one of the National Institute of Allergy and Infectious Diseases (NIAID) funded Centers of Excellence for Influenza Research and Response (CEIRR; contract # 75N93021C00014); the San Antonio Partnership for Precision Therapeutics; the Texas Biomedical Research Institute Forum Awards; and the San Antonio Medical Foundation. Research in LM-S was also partially supported by NIH R01AI145332, R01AI142985, and R01AI141607; the DoD W81XWH1910496; and by the American Lung Association.
H.D. conceptualized the project and performed the vaccination studies including antibody and T-cell responses to the NP/RBD protein. K.W. helped clone the genes. A.N. and J.C. helped perform the mouse vaccinations. A.N. and C.C. helped collect sera and spleen samples from immunized mice. D.X. performed part of the SARS-CoV-2 neutralization assays. E.V. and E.H.W. helped with immune analysis. R.B., M.S.Y., S.R.K., and L.M.S. performed in vivo infection studies. A.U. and N.B. helped perform the CD4 T-cell depletion and associated vaccination studies. R.H. designed, supervised, and organized the study. H.D. and R.H. wrote the manuscript with input from other authors.
Contributor Information
Rong Hai, Email: ronghai@ucr.edu.
Mark T. Heise, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina, USA
DATA AVAILABILITY
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplemental material.
ETHICS APPROVAL
Animal studies were approved by the University of California, Riverside, Institutional Animal Care and Use Committee (IACUC) and performed in the biosafety level 2 facility. Neutralization assays with SARS-CoV-2 virus were performed in the UCR biosafety level 3 facility. All animal SARS-CoV-2 virus challenging studies were reviewed and approved by the Texas Biomedical Research Institute (Texas Biomed) Animal Care and Use Committee (IACUC) and conducted at ABSL3 containment. All animals were cared for in the Animal Resources Facility under specific-pathogen-free conditions in compliance with the Institute for Laboratory Animal Research Guide for the Care and Use of Laboratory Animals, 8th edition. Both the University of California, Riverside, Institutional Biosafety Committee (IBC) and Texas Biomed IBC approved the procedures for sample handling, inactivation, and removal from the corresponding BSL3 containment.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/jvi.01571-23.
Fig. S1 to S4; Table S1.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Stadler K, Masignani V, Eickmann M, Becker S, Abrignani S, Klenk H-D, Rappuoli R. 2003. SARS--beginning to understand a new virus. Nat Rev Microbiol 1:209–218. doi: 10.1038/nrmicro775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Raj VS, Osterhaus A, Fouchier RAM, Haagmans BL. 2014. MERS: emergence of a novel human coronavirus. Curr Opin Virol 5:58–62. doi: 10.1016/j.coviro.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. Author correction: a new coronavirus associated with human respiratory disease in China. Nature 580:E7. doi: 10.1038/s41586-020-2202-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, et al. 2020. An mRNA vaccine against SARS-CoV-2- preliminary report. N Engl J Med 383:1920–1931. doi: 10.1056/NEJMoa2022483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Raabe V, Bailey R, Swanson KA, et al. 2020. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586:589–593. doi: 10.1038/s41586-020-2639-4 [DOI] [PubMed] [Google Scholar]
- 6. Tregoning JS, Flight KE, Higham SL, Wang Z, Pierce BF. 2021. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat Rev Immunol 21:626–636. doi: 10.1038/s41577-021-00592-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tian D, Sun Y, Xu H, Ye Q. 2022. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 omicron variant. J Med Virol 94:2376–2383. doi: 10.1002/jmv.27643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky Pond SL, Fera D, Shafer RW. 2021. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet 22:757–773. doi: 10.1038/s41576-021-00408-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Consortium C-G, Peacock SJ, Robertson DL. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. doi: 10.1038/s41579-021-00573-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yu J, Collier A-R, Rowe M, Mardas F, Ventura JD, Wan H, Miller J, Powers O, Chung B, Siamatu M, Hachmann NP, Surve N, Nampanya F, Chandrashekar A, Barouch DH. 2022. Neutralization of the SARS-CoV-2 omicron BA.1 and BA.2 variants. N Engl J Med 386:1579–1580. doi: 10.1056/NEJMc2201849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, Rangarajan ES, Pan A, Vanderheiden A, Suthar MS, Li W, Izard T, Rader C, Farzan M, Choe H. 2020. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun 11:6013. doi: 10.1038/s41467-020-19808-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou B, Thao TTN, Hoffmann D, Taddeo A, Ebert N, Labroussaa F, Pohlmann A, King J, Steiner S, Kelly JN, et al. 2021. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 592:122–127. doi: 10.1038/s41586-021-03361-1 [DOI] [PubMed] [Google Scholar]
- 13. Milbank C, Vira B. 2022. Wildmeat consumption and zoonotic spillover: contextualising disease emergence and policy responses. Lancet Planet Health 6:e439–e448. doi: 10.1016/S2542-5196(22)00064-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tajudeen YA, Oladunjoye IO, Bajinka O, Oladipo HJ. 2022. Zoonotic spillover in an era of rapid deforestation of tropical areas and unprecedented wildlife trafficking: into the wild. Challenges 13:41. doi: 10.3390/challe13020041 [DOI] [Google Scholar]
- 15. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 183:1735. doi: 10.1016/j.cell.2020.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yurkovetskiy L, Wang X, Pascal KE, Tomkins-Tinch C, Nyalile TP, Wang Y, Baum A, Diehl WE, Dauphin A, Carbone C, et al. 2020. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 183:739–751. doi: 10.1016/j.cell.2020.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cai Y, Zhang J, Xiao T, Peng H, Sterling SM, Walsh RM, Rawson S, Rits-Volloch S, Chen B. 2020. Distinct conformational States of SARS-CoV-2 spike protein. Science 369:1586–1592. doi: 10.1126/science.abd4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ace2 receptor. Nature 581:215–220. doi: 10.1038/s41586-020-2180-5 [DOI] [PubMed] [Google Scholar]
- 19. Alleva DG, Delpero AR, Scully MM, Murikipudi S, Ragupathy R, Greaves EK, Sathiyaseelan T, Haworth JR, Shah NJ, Rao V, Nagre S, Lancaster TM, Webb SS, Jasa AI, Ronca SE, Green FM, Elyard HA, Yee J, Klein J, Karnes L, Sollie F, Zion TC. 2021. Development of an IgG-FC fusion COVID-19 subunit vaccine, AKS-452. Vaccine 39:6601–6613. doi: 10.1016/j.vaccine.2021.09.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. An Y, Li S, Jin X, Han J-B, Xu K, Xu S, Han Y, Liu C, Zheng T, Liu M, et al. 2022. A tandem-repeat dimeric RBD protein-based COVID-19 vaccine zf2001 protects mice and nonhuman primates. Emerg Microbes Infect 11:1058–1071. doi: 10.1080/22221751.2022.2056524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dai L, Gao L, Tao L, Hadinegoro SR, Erkin M, Ying Z, He P, Girsang RT, Vergara H, Akram J, et al. , Group ZFGT . 2022. Efficacy and safety of the RBD-dimer-based COVID-19 vaccine ZF2001 in adults. N Engl J Med 386:2097–2111. doi: 10.1056/NEJMoa2202261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Z, Xu W, Xia S, Gu C, Wang X, Wang Q, Zhou J, Wu Y, Cai X, Qu D, Ying T, Xie Y, Lu L, Yuan Z, Jiang S. 2020. RBD-FC-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct Target Ther 5:282. doi: 10.1038/s41392-020-00402-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ma X, Zou F, Yu F, Li R, Yuan Y, Zhang Y, Zhang X, Deng J, Chen T, Song Z, Qiao Y, Zhan Y, Liu J, Zhang J, Zhang X, Peng Z, Li Y, Lin Y, Liang L, Wang G, Chen Y, Chen Q, Pan T, He X, Zhang H. 2020. Nanoparticle vaccines based on the receptor binding domain (RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. Immunity 53:1315–1330. doi: 10.1016/j.immuni.2020.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun S, Cai Y, Song TZ, Pu Y, Cheng L, Xu H, Sun J, Meng C, Lin Y, Huang H, et al. 2021. Interferon-armed RBD dimer enhances the immunogenicity of RBD for sterilizing immunity against SARS-CoV-2. Cell Res 31:1011–1023. doi: 10.1038/s41422-021-00531-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tai W, Zhang X, Drelich A, Shi J, Hsu JC, Luchsinger L, Hillyer CD, Tseng C-TK, Jiang S, Du L. 2020. A novel receptor-binding domain (RBD)-based mRNA vaccine against SARS-CoV-2. Cell Res 30:932–935. doi: 10.1038/s41422-020-0387-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang J, Wang W, Chen Z, Lu S, Yang F, Bi Z, Bao L, Mo F, Li X, Huang Y, et al. 2020. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586:572–577. doi: 10.1038/s41586-020-2599-8 [DOI] [PubMed] [Google Scholar]
- 27. Yang S, Li Y, Dai L, Wang J, He P, Li C, Fang X, Wang C, Zhao X, Huang E, et al. 2021. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect Dis 21:1107–1119. doi: 10.1016/S1473-3099(21)00127-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mamatkulov M. 2021. Uzbekistan APPROVES Chinese-developed COVID-19 vaccine. Reuters
- 29. Anonymous . 2021. China IMCAS’s COVID-19 vaccine obtained emergency use approval in China. Reuters
- 30. Perappadan BS. 2022. Corbevax, covaxin get drug regulator nod for use in children between 5 and 12, on The Hindu. Available from: https://www.thehindu.com/sci-tech/health/covid-19-vaccines-corbevax-covaxin-get-drug-regulator-nod-for-use-in-children-between-5-and-12/article65356421.ece
- 31. Pegu A, O’Connell SE, Schmidt SD, O’Dell S, Talana CA, Lai L, Albert J, Anderson E, Bennett H, Corbett KS, et al. 2021. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science 373:1372–1377. doi: 10.1126/science.abj4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walsh EE, Frenck RW, Falsey AR, Kitchin N, Absalon J, Gurtman A, Lockhart S, Neuzil K, Mulligan MJ, Bailey R, et al. 2020. Safety and immunogenicity of two RNA-based COVID-19 vaccine candidates. N Engl J Med 383:2439–2450. doi: 10.1056/NEJMoa2027906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, Flach B, O’Connell S, Bock KW, Minai M, et al. 2020. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med 383:1544–1555. doi: 10.1056/NEJMoa2024671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abu-Raddad LJ, Chemaitelly H, Bertollini R, National Study Group for COVID-19 Vaccination . 2022. Waning mRNA-1273 vaccine effectiveness against SARS-CoV-2 infection in Qatar. N Engl J Med 386:1091–1093. doi: 10.1056/NEJMc2119432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lopez Bernal J, Gower C, Andrews N. 2021. Effectiveness of COVID-19 vaccines against the B.1.617.2 (Delta) variant. reply. N Engl J Med 385:e92. doi: 10.1056/NEJMc2113090 [DOI] [PubMed] [Google Scholar]
- 36. DiPiazza AT, Fan S, Rattan A, DeDiego ML, Chaves F, Neumann G, Kawaoka Y, Sant AJ. 2019. A novel vaccine strategy to overcome poor immunogenicity of avian influenza vaccines through mobilization of memory CD4 T cells established by seasonal influenza. J Immunol 203:1502–1508. doi: 10.4049/jimmunol.1900819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gallagher JR, Torian U, McCraw DM, Harris AK. 2017. Structural studies of influenza virus RNPs by electron microscopy indicate molecular contortions within NP supra-structures. Journal of Structural Biology 197:294–307. doi: 10.1016/j.jsb.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with alphafold. Nature 596:583–589. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chiem K, Park JG, Morales Vasquez D, Plemper RK, Torrelles JB, Kobie JJ, Walter MR, Ye C, Martinez-Sobrido L. 2022. Monitoring SARS-CoV-2 infection using a double reporter-expressing virus. Microbiol Spectr 10:e0237922. doi: 10.1128/spectrum.02379-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zang J, Gu C, Zhou B, Zhang C, Yang Y, Xu S, Bai L, Zhang R, Deng Q, Yuan Z, Tang H, Qu D, Lavillette D, Xie Y, Huang Z. 2020. Immunization with the receptor-binding domain of SARS-CoV-2 elicits antibodies cross-neutralizing SARS-CoV-2 and SARS-CoV without antibody-dependent enhancement. Cell Discov 6:61. doi: 10.1038/s41421-020-00199-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ravichandran S, Coyle EM, Klenow L, Tang J, Grubbs G, Liu S, Wang T, Golding H, Khurana S. 2020. Antibody signature induced by SARS-CoV-2 spike protein immunogens in rabbits. Sci Transl Med 12:eabc3539. doi: 10.1126/scitranslmed.abc3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shajahan A, Supekar NT, Gleinich AS, Azadi P. 2020. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 30:981–988. doi: 10.1093/glycob/cwaa042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. 2020. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369:330–333. doi: 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Guthmiller JJ, Utset HA, Henry C, Li L, Zheng N-Y, Sun W, Costa Vieira M, Zost S, Huang M, Hensley SE, Cobey S, Palese P, Wilson PC. 2021. An egg-derived sulfated N-acetyllactosamine glycan is an antigenic decoy of influenza virus vaccines. mBio 12:e0083821. doi: 10.1128/mBio.00838-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brown EL, Essigmann HT, Pascual DW. 2021. Original antigenic sin: the downside of immunological memory and implications for COVID-19. mSphere 6:e00056-21. doi: 10.1128/mSphere.00056-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. East IJ, Todd PE, Leach SJ. 1980. Original Antigenic sin: experiments with a defined antigen. Mol Immunol 17:1539–1544. doi: 10.1016/0161-5890(80)90179-0 [DOI] [PubMed] [Google Scholar]
- 47. Vatti A, Monsalve DM, Pacheco Y, Chang C, Anaya JM, Gershwin ME. 2017. Original antigenic sin: a comprehensive review. J Autoimmun 83:12–21. doi: 10.1016/j.jaut.2017.04.008 [DOI] [PubMed] [Google Scholar]
- 48. Zhang A, Stacey HD, Mullarkey CE, Miller MS. 2019. Original antigenic sin: how first exposure shapes lifelong anti-influenza virus immune responses. J Immunol 202:335–340. doi: 10.4049/jimmunol.1801149 [DOI] [PubMed] [Google Scholar]
- 49. Doan TA, Forward T, Tamburini BAJ. 2022. Trafficking and retention of protein antigens across systems and immune cell types. Cell Mol Life Sci 79:275. doi: 10.1007/s00018-022-04303-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rodrigues CMC, Plotkin SA. 2021. The influence of interval between doses on response to vaccines. Vaccine 39:7123–7127. doi: 10.1016/j.vaccine.2021.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dedroogh S, Schmiedl S, Thürmann PA, Graf K, Appelbaum S, Koß R, Theis C, Zia Z, Tebbenjohanns J, Thal SC, Dedroogh M. 2023. Impact of timing and combination of differentt BNT162b2 and ChAdOx1-S COVID-19 basic and booster vaccinations on humoral immunogenicity and reactogenicity in adults. Sci Rep 13:9036. doi: 10.1038/s41598-023-34961-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Petrie JG, Ohmit SE, Johnson E, Truscon R, Monto AS. 2015. Persistence of antibodies to influenza hemagglutinin and neuraminidase following one or two years of influenza vaccination. J Infect Dis 212:1914–1922. doi: 10.1093/infdis/jiv313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dai L, Zheng T, Xu K, Han Y, Xu L, Huang E, An Y, Cheng Y, Li S, Liu M, Yang M, Li Y, Cheng H, Yuan Y, Zhang W, Ke C, Wong G, Qi J, Qin C, Yan J, Gao GF. 2020. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell 182:722–733. doi: 10.1016/j.cell.2020.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lell B, Agnandji S, von Glasenapp I, Haertle S, Oyakhiromen S, Issifou S, Vekemans J, Leach A, Lievens M, Dubois M-C, Demoitie M-A, Carter T, Villafana T, Ballou WR, Cohen J, Kremsner PG. 2009. A randomized trial assessing the safety and immunogenicity of AS01 and AS02 adjuvanted RTS,S malaria vaccine candidates in children in gabon. PLOS ONE 4:e7611. doi: 10.1371/journal.pone.0007611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kingston NJ, Walsh R, Hammond R, Joe CCD, Lovrecz G, Locarnini S, Netter HJ. 2023. Immunogenicity of wild type and mutant hepatitis B surface antigen virus-like particles (VLPs) in mice with pre-existing immunity against the wild type vector. Viruses 15:313. doi: 10.3390/v15020313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hoffmann AB, Mazelier M, Léger P, Lozach P-Y. 2018. Deciphering virus entry with fluorescently labeled viral particles. Methods Mol Biol 1836:159–183. doi: 10.1007/978-1-4939-8678-1_8 [DOI] [PubMed] [Google Scholar]
- 57. Ye C, Chiem K, Park J-G, Silvas JA, Morales Vasquez D, Sourimant J, Lin MJ, Greninger AL, Plemper RK, Torrelles JB, Kobie JJ, Walter MR, de la Torre JC, Martinez-Sobrido L. 2021. Analysis of SARS-CoV-2 infection dynamic in vivo using reporter-expressing viruses. Proc Natl Acad Sci USA 118. doi: 10.1073/pnas.2111593118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mendoza EJ, Manguiat K, Wood H, Drebot M. 2020. Two detailed plaque assay protocols for the quantification of infectious SARS-CoV-2. Curr Protoc Microbiol 57:ecpmc105. doi: 10.1002/cpmc.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408 [DOI] [Google Scholar]
- 60. Ayasoufi K, Fan R, Valujskikh A. 2017. Depletion-resistant CD4 T cells enhance thymopoiesis during lymphopenia. Am J Transplant 17:2008–2019. doi: 10.1111/ajt.14309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4; Table S1.
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or the supplemental material.