Fig 4.
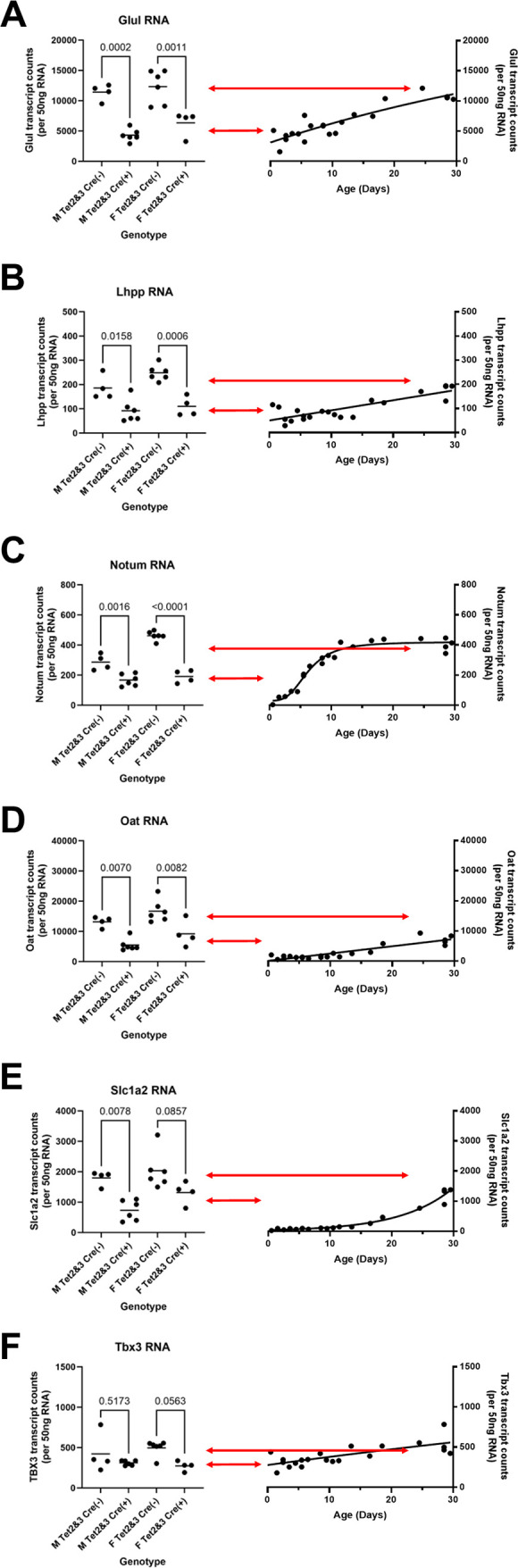
Effects of Tet deletion on liver β-catenin target gene transcript levels in HBV transgenic mice. Quantitative analysis of (A) Glul, (B) Lhpp, (C) Notum, (D) Oat, (E) Slc1a2, and (F) Tbx3 transcripts by NanoString gene expression analysis in HBV transgenic mice. Individual (circles) and mean (line) transcript levels are indicated for male (M), female (F), homozygous Tet2(fl/fl)Tet3(fl/fl) alleles (Tet2 and Tet3), Cre-negative allele [Cre(−)], and Cre-positive allele [Cre(+)] HBV transgenic mice. Statistical differences (adjusted P values; Tukey’s multiple comparison test) in the levels of the transcripts between Cre(−) and Cre(+) HBV transgenic mice are indicated (GraphPad Prism 10.0.3). Transcript levels from adult Tet-deficient HBV transgenic mice (left panel) are compared to the changes in gene expression observed throughout postnatal liver development in wild-type HBV transgenic mice (right panel). Arrows indicate the average levels of transcript observed in HBVTet2(fl/fl)Tet3(fl/fl)Cre(−) (top arrow) and HBVTet2(fl/fl)Tet3(fl/fl)Cre(+) (bottom arrow) mice.
