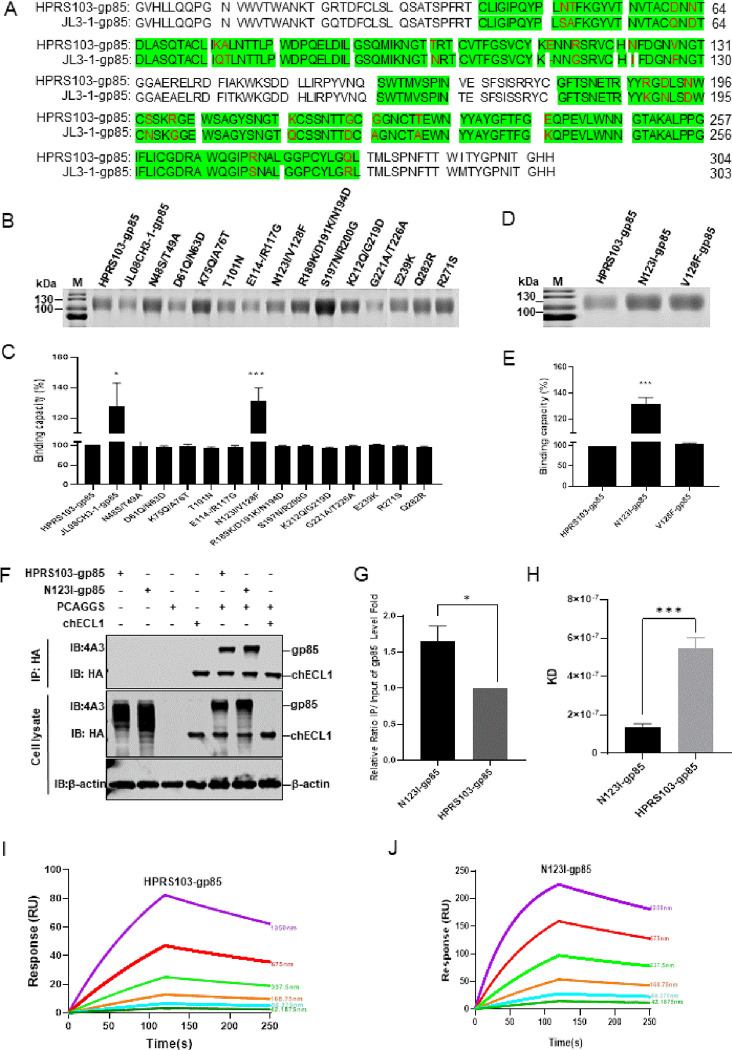
Fig 3. Increased binding of the N123I protein to the receptor.
(A) Amino acid sequence alignment analysis for gp85 from HPRS103 and JL08CH3-1. Green shading represents the chNHE1-binding domain of ALV-J-gp85. The amino acid mutations within the RBD region are marked in red. (B) SDS-PAGE analysis of the JL08CH3-1-gp85, HPRS103-gp85, and 13 mutation gp85 proteins (according to the previous approach to construction). Fifteen plasmids containing CAG-sgp85-Fc, CAG-JL3-1-p85-Fc, and 13 mutation gp85 proteins are transfected into 293T cells for 48 h, then harvested supernatant of cells is purified by using Protein A Resin. Subsequently, the purified gp85 proteins are verified by SDS-PAGE analysis. (C) Binding capacity between the JL08CH3-1-gp85, HPRS103-gp85, and 13 mutation gp85 proteins with chNHE1 expressed on the surface of transfected 293T cells. (D) SDS-PAGE analysis of the N123I-gp85, V128F-gp85, and HPRS103-gp85 proteins. The CAG-sgp85-Fc, CAG-N123I-gp85-Fc, and CAG-V128F-gp85-Fc plasmids are transfected into 293T cells for 48 h, then harvested supernatant of cells is purified using Protein A Resin. Subsequently, the purified gp85 proteins are verified using SDS-PAGE analysis. (E) Binding capacity between the N123I-gp85, V128F-gp85, and HPRS103-gp85 proteins with chNHE1 expressed on the surface of transfected 293T cells. (F and G) The binding capacity is detected by Co-IP assays in 293T cells. (F) 293T cells are co-transfected with chECL1 and pCAF-N123I-gp85 or pCAF-gp85 for 48 h. The lysates are incubated with anti-HA-agarose MAb. Then, the lysates are detected by 4A3 monoclonal antibody and anti-HA monoclonal antibody using Western blotting. (G) Relative intensities of gp85 are normalized to the gp85 in the input sample. (H) Statistical summary of the KD values of N123I-gp85 or HPRS103-gp85 and chECL1. (I and J) The binding kinetics of the chECL1 proteins with HPRS103-gp85(I) or N123I-gp85 (J) are detected using the Biacore 8K (Cytiva, Marlborough, MA, USA). ChECL1 protein is captured on the chip, and serial dilutions of gp85 then flow through the chip surface. Experiments are performed three times, revealing similar results, and one set of representative data is displayed. The error bars represent the SD. *, P < 0.05; **, P < 0.01; *** and ****, P < 0.001; ns, not significant; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; RBD, receptor-binding domain; KD, equilibrium dissociation constant; SD, standard deviation; Co-IP, coimmunoprecipitation; chECL1, chicken first extracellular loop of chNHE1; chNHE1, chicken sodium hydrogen exchanger isoform 1.