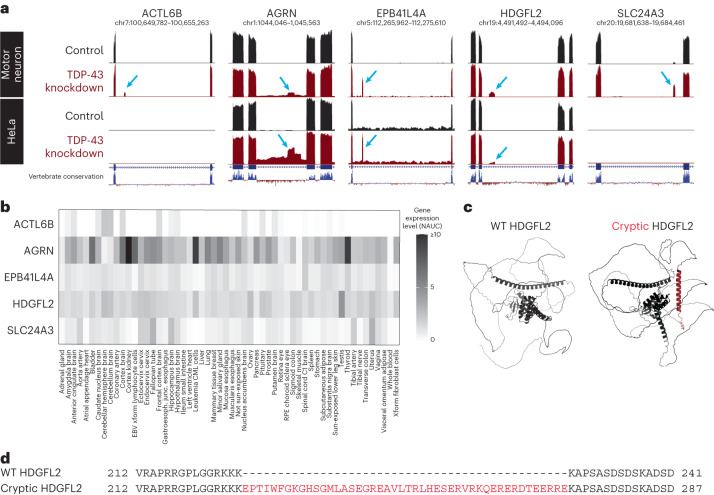
Fig. 1. Identification of human in-frame, TDP-43-associated cryptic exons.
a, UCSC Genome Browser visualization of selected cryptic exons in human motor neurons11 and HeLa cells7 aligned to the GRCh38 assembly. Red tracks indicate TDP-43 knockdown, and blue arrows identify nonconserved cryptic exons. Gene annotations below RNA sequencing tracks indicate canonical exons (thick lines) and introns (thin lines). b, Visualization of tissue type-specific gene expression of ACTL6B, AGRN, EPB41L4A, HDGFL2 and SLC24A3. While AGRN and HDGFL2 RNA transcripts are ubiquitously expressed, ACTL6B, EPB41L4A and SLC24A3 are expressed in a more tissue-specific manner. NAUC values derived from ASCOT19 are used to approximate gene expression levels in different human tissue types. c, Comparison of WT (left) and cryptic (right) HDGFL2 protein structures. Inclusion of the cryptic exon in mRNA leads to the addition of 46 amino acids predicted to form an alpha-helix structure (red) between flanking unstructured regions. Both structures are generated using AlphaFold predictions derived from amino acid sequences. The WT HDGFL2 protein structure can be found on the AlphaFold protein structure database (UniProt: Q7Z4V5). d, Alignment of WT and cryptic HDGFL2 amino acid sequences. The cryptic inclusion is 46 amino acids long (red) and does not impact flanking amino acids.