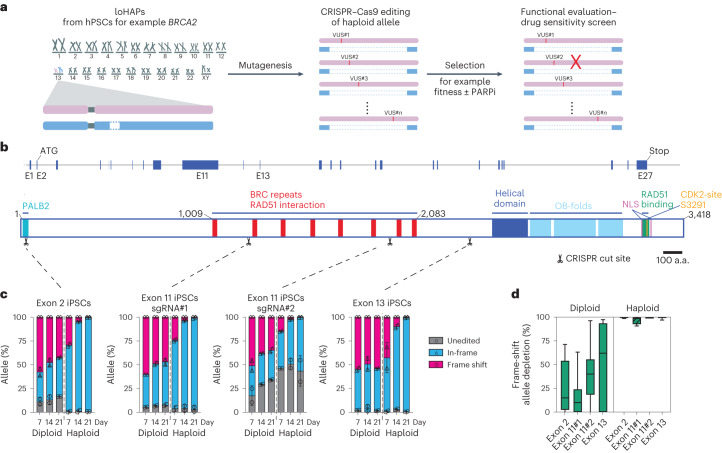
Fig. 1. Annotating functional protein domains in BRCA2 using CRISPR-mediated mutagenesis in BRCA2 loHAPs.
a, Annotation of genetic variants using loHAPs. First, one copy of a gene of interest (such as BRCA2) is excised from hPSCs to generate loHAPs. In a second step, the remaining allele is mutagenized (by introducing VUS) to identify alleles that are permissive and stably maintained in the cell pool. Finally, the pool of cells is challenged (for example, with a PARP inhibitor in BRCA2 loHAPs) to identify variants sensitive to the treatment. b, Schematic representation of the BRCA2 locus and the BRCA2 protein domain structure. Shown are the BRCA2 exons (blue boxes), the sgRNA target sites used in this study (indicated with scissors) as well as the BRCA2 protein domain structure: PALB2 domain (turquoise), BRC repeats (red), helical domain (dark blue), OB-folds (light blue), C-terminal RAD51 binding domain (green), two nuclear localization signals (NLS, purple) and CDK2 phosphorylation sites (orange). Scale bar, 100 amino acids (a.a.). c, Quantification of categorized allele frequencies (unedited, in-frame and frame-shift indels) in exons 2, 11 and 13 assayed in diploid and BRCA2 loHAP hiPSCs at days 7, 14 and 21 after editing. Bars showing mean of two biological replicates and error bars showing s.e.m. d, Quantification of the relative depletion of frame-shift mutation in exons 2, 11 and 13 comparing samples collected at day 7 and 21 after editing in diploid and BRCA2 loHAP hiPSCs. Boxes showing quartiles and whiskers showing the 10th and 90th percentile. Mutations with frequency >0.5% are plotted.