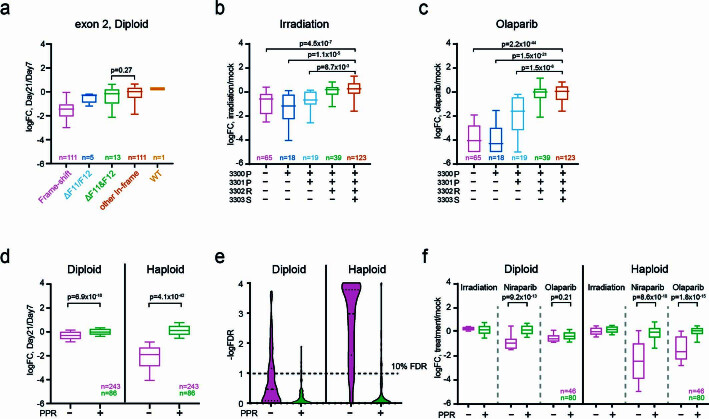
Extended Data Fig. 6. Comparison between diploid and BRCA2 loHAPs in identifying functionally critical amino acids by CRISPR-mediated mutation tiling.
a, Categorized allele frequency changes of F11, F12 deletions at day 21 comparing to day 7, presented as log2(day 21/day 7) in diploid. P value reports the significance between F11& F12 double deletion vs. other in-frame mutations in two-tailed Student’s t-test. n shown number of different alleles in each group. Boxes showing quartiles and Whiskers showing the 10th and 90th percentile. b, Categorized allele frequency changes of frame-shift mutations break before P3300, at P3300, at P3301, at R3302 or at later positions, upon irradiation comparing to the mock condition, presented as log2(irradiation/mock). Boxes showing quartiles and Whiskers showing the 10th and 90th percentile. P, FDR-corrected two-tailed student’s t-test vs. the PPRS fully preserved group. N, number of different alleles in each group. c, Same as b, for Olaparib treatment. d, Categorized allele frequency changes of frame-shift mutations with PPR motif preserved (+) or destroyed (−) assayed in diploid and BRCA2 loHAP hESCs in an independent tiling deletion screen, at day 21 comparing to day 7, presented as log2(day 21/day 7). Boxes showing quartiles and Whiskers showing the 10th and 90th percentile. P, two-tailed student’s t-test. n, number of different alleles in each group. e, The distribution of FDR-corrected p value of each allele shown in d. The p values of alleles that frequencies increased, were assigned to be 1 (-logFDR=0). f, Same as d, upon treatment of irradiation, Niraparib or Olaparib comparing to the mock condition, presented as log2(treatment/mock).