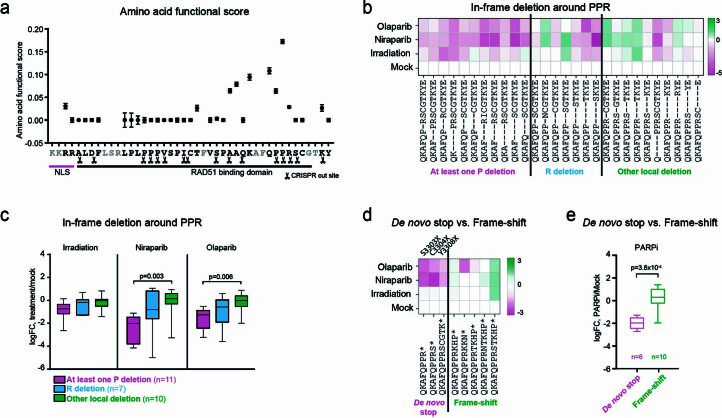
Extended Data Fig. 7. Evaluating the function of critical amino acids by analyzing in-frame deletions generated in CRISPR-mediated mutation tiling.
a, Functional score of amino acid in the BRCA2 RAD51 binding domain encoded in exon 27 calculated by a statistical model, with CRISPR cut site labeled. Only scores of amino acids covered by one amino acid deletions are plotted. b, The aligned allele profile of <=4 amino acids in-frame deletions around PPR (3300–3302) and heatmaps of allele frequency changes upon the treatment of irradiation or PARPi. Alleles were grouped by the amino acid in PPR motif deleted. Allele frequencies of each mutation were normalized to the corresponding allele frequencies in mock. c, Categorized allele frequency changes of alleles in b. Boxes showing quartiles and Whiskers showing the 10th and 90th percentile. p, FDR-corrected two-tailed student’s t-test. n, number of different alleles in each group. d, The aligned allele profile of de novo stops and frame-shift mutation that result in stop codons between S3303 and Y3308 and heatmaps of allele frequency changes upon the treatment of irradiation or PARPi as shown in b. Note: We identified three de novo stop mutations S3303X, C3304X and Y3308X. In each case these de novo stops depleted after PARPi suggesting that de novo stop codons between 3302 and 3308 have reduced fitness suggesting that the presence of amino acids (even it not generated by the BRCA2 frame) between amino acid 3302 and 3308 contributes to BRCA2 protein function. These findings are consistent with a pathogenic de novo stop codon identified at amino acid 3308. e, Categorized allele frequency changes of alleles in d upon PARP inhibition. p, two-tailed student’s t-test. n, number of measurements in each group. The responses of each allele to Niraparib or Olaparib were considered as separate measurements.