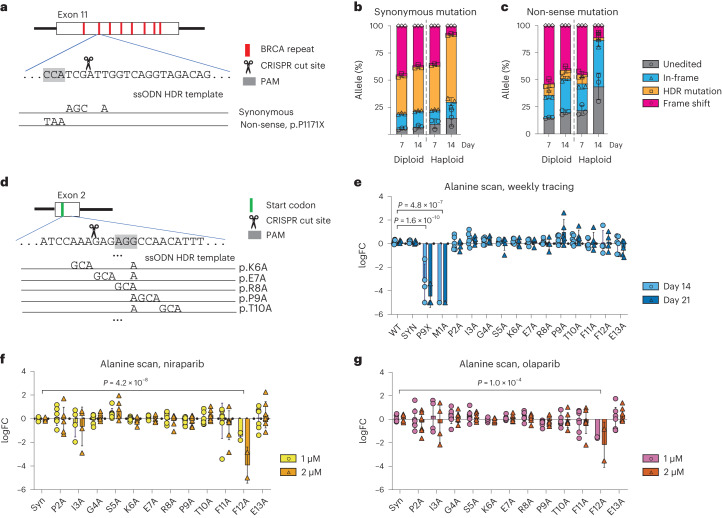
Fig. 3. Using HDR-mediated genome engineering in BRCA2 loHAPs to assess functionality of designer mutations.
a, HDR-based editing strategy to introduce designer mutations in exon 11 of BRCA2. Shown is the targeted sequence with the PAM shaded in grey. Indicated below are the two HDR single-stranded repair oligos specifying the mutations that result in either synonymous or non-sense mutations. b, Quantification of categorized allele frequencies (unedited, in-frame indels, HDR-synonymous mutation and frame-shift mutations) in exon 11 assayed in diploid and BRCA2 loHAP hiPSCs at days 7 and 14 after editing. Bars showing mean of three biological replicates and error bars showing s.e.m. c, Analysis as in b, using the HDR template introducing a non-sense mutation. d, HDR-based editing strategy to perform alanine scanning in BRCA2. Shown is the targeted sequence within exon 2 with the PAM shaded in grey. Indicated below are examples for the HDR single-stranded repair oligos, specifying the mutations that result in alanine replacements. The start codon is indicated in green. e, Allele frequency changes over time of mutations generated in BRCA2 exon 2 alanine scanning in BRCA2 loHAP hPSCs, represented as log2(day 14/day 7), circle, and log2(day 21/day 7), triangle. P, FDR-corrected two-tailed Student’s t-test versus WT. Bars showing mean and error bars showing s.e.m. Data points represent biological replicates. FC, fold change. f, Normalized allele frequency changes upon treatment of PARP inhibitor, niraparib, of BRCA2 exon 2 alanine mutations in BRCA2 loHAP hPSCs, represented as log2(drug/mock), 1 µM, circle; 2 µM, triangle. P, FDR-corrected two-tailed Student’s t-test versus synonymous mutations (Syn). Allele frequencies were normalized against corresponding WT. g, Same as in f, for olaparib treatment.