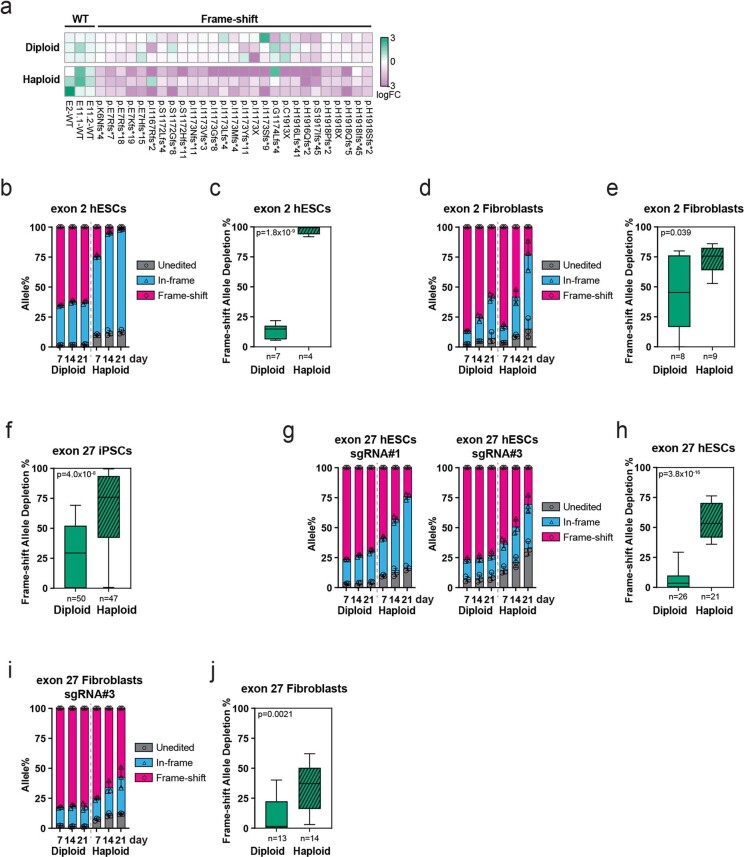
Extended Data Fig. 2. Annotating BRCA2 protein domain using CRISPR-mediated mutagenesis in loHAP hiPSCs, hESCs and hPSCs-derived fibroblasts.
a, Heatmap indicating the allele frequency changes, log2 (day 14/day 7), of individual frame-shift mutation in 3 biological replicates in hiPSCs. b, Quantification of categorized allele frequencies (unedited, in-frame and frame-shift indels) for an sgRNA targeting exon 2 assayed in diploid and BRCA2 loHAP hESCs at day 7, 14 and 21 after editing. Bars showing mean of 3 biological replicates and error bars showing SEM. c, Quantification of the relative depletion of frame-shift mutation in exon 2 comparing samples collected at day 7 and 21 after editing in diploid and BRCA2 loHAP hESCs. Boxes showing quartiles and whiskers showing the 10th and 90th percentile. Mutations with frequency >0.5% are plotted. p, two-tailed Student’s t-test; n, number of alleles. d, Same as in b, for a sgRNA targeting exon 2 in hPSCs derived fibroblasts. e, Same as in c, for alleles generated in exon 2 in hPSCs derived fibroblasts. f, Same as in c, for alleles generated in exon 27 in hiPSCs. g, Same as in b, for sgRNAs targeting exon 27 in hESCs. h, Same as in c, for alleles generated in exon 27 in hESCs. i, Same as in b. for a sgRNA targeting exon 27 in hPSCs derived fibroblasts. j, Same as in c, for alleles generated in exon 27 in hPSCs derived fibroblasts.