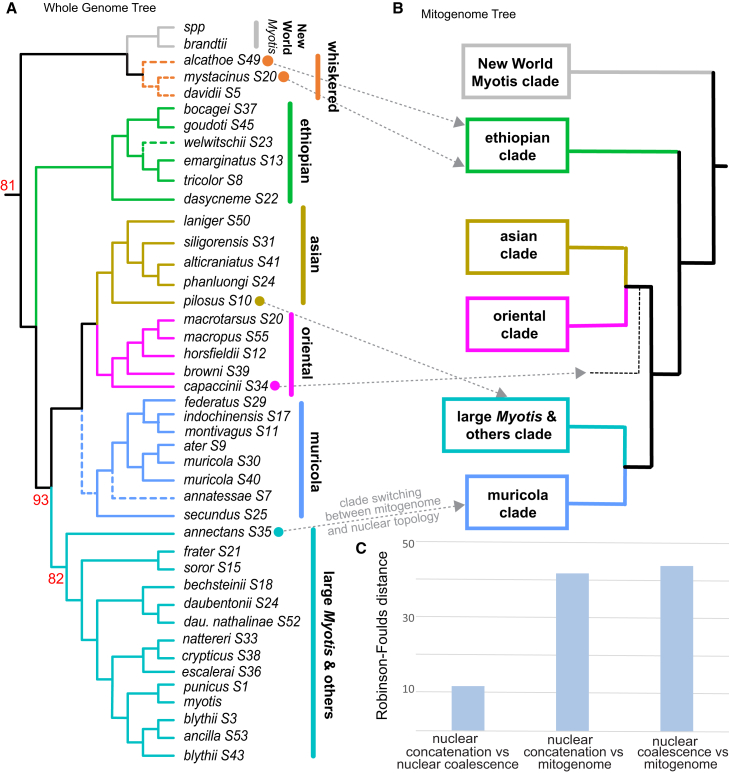
Figure 1.
Clade switching between the whole-genome nuclear and mitogenome topologies
(A) Maximum likelihood cladogram from ∼1.5 million genome-wide SNVs, showing only one sample per species where multiple exist. Bootstrap support is 100% for all nodes unless indicated otherwise. Dashed branches show which relationships differ between concatenation and coalescent analyses of the same dataset. Concatenation bootstrap values are shown in red.
(B) The mitogenome phylogeny is illustrated to highlight, via gray dashed arrows, clade switching relative to the nuclear phylogeny for the species indicated with colored dots.
(C) Robinson-Foulds (RF) distances between the nuclear and mitogenome topologies.