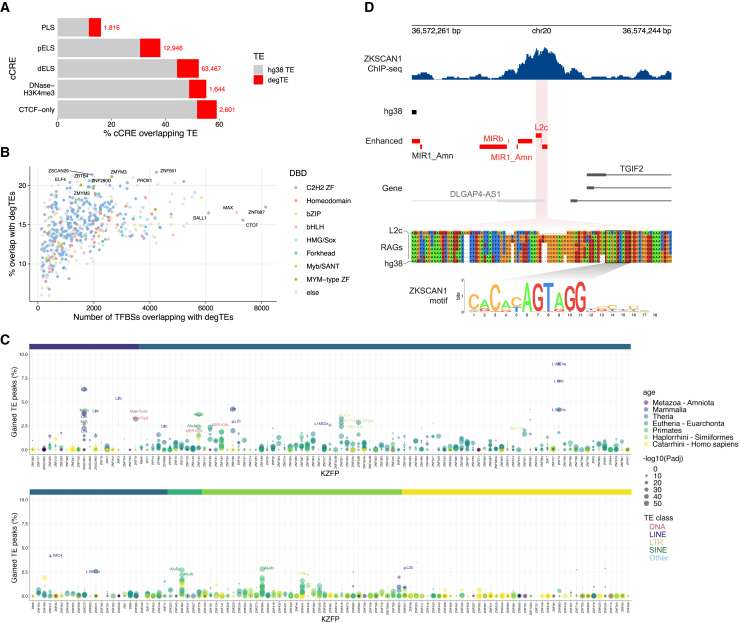
Figure 4.
cCREs and TFBSs derived from degTEs
(A) Percentage of cCREs overlapping the hg38 TE and degTEs. The red numbers indicate cCREs that overlap with one or more degTEs. PLS, promoter-like signatures; pELS, proximal enhancer-like signatures; dELS, distal enhancer-like signatures.
(B) ENCODE TFBSs overlapping with one or more degTEs. The colors represent DNA-binding domains of the TFs.
(C) A summary of degTE-overlapping KZFP binding sites shown in percentage relative to the total number of peaks. Each point represents a TE subfamily and its color and size indicate its evolutionary age and an adjusted enrichment p-value, respectively. The same color scale is used for the ribbon above to indicate the ages of the KZFPs.
(D) A representative genomic region where a degTE-derived KZFP binding site was found. A multiple sequence alignment is shown for the newly found L2c element in the middle of the ZKSCAN1 peak and its corresponding sequences in the RAGs together with the L2c consensus. The sequence matching the ZKSCAN1 binding motif is highlighted.