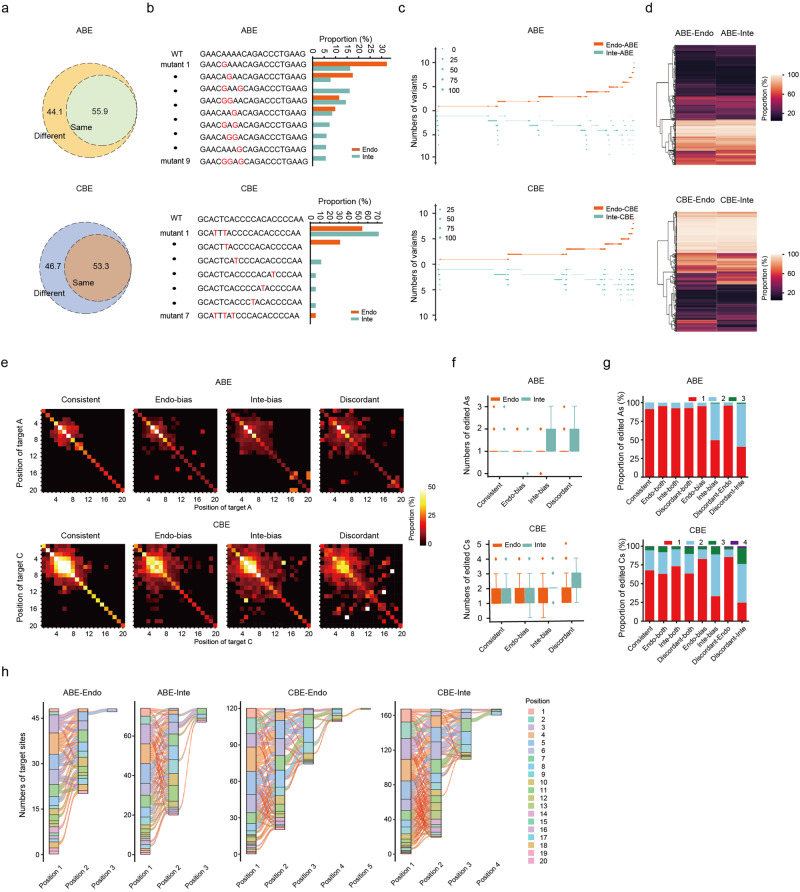
Fig. 4. Comparison of editing outcomes of ABE and CBE between endogenous and integrated target sites.
a Comparison of the editing outcome alleles between the Endo- and Inte- datasets for ABE and CBE. b Representative editing outcomes of ABE or CBEs at endogenous and integrated target sites with the same sequence. c Comparison of the editing outcome products between the Endo- and Inte-datasets for ABE and CBE. The dot size represents percentage of unique editing outcome products in Endo- or Inte- dataset. The target sites were thus divided into 4 groups: “Consistent”, target sites with consistent editing outcomes between the Endo- and Inte- datasets; “Endo-bias” and “Inte-bias” groups, target sites with specific alleles in the Endo- or Inte- datasets, respectively; “Discordant” group, target sites with specific alleles in both Endo- and Inte-datasets. d Comparison of the proportions of same editing outcome products in the consistent group of ABE and CBE. e Co-occurrence proportions of each targeted As or Cs. f Comparison of the number of edited As (ABE) or Cs (CBE) within each target site among the “Consistent”, “Endo-bias”, “Inte-bias”, and “Discordant” groups of ABE or CBE. n = 2005 (Consistent), 824 (Endo-bias), 511 (Inte-bias) and 245 (Discordant) for ABE. n = 1930 (Consistent), 877 (Endo-bias), 552 (Inte-bias) and 263 (Discordant) for ABE. g Normalized proportion of edited As (ABE) or Cs (CBE) within each target site among the “Consistent”, “Endo-bias”, “Inte-bias”, and “Discordant” groups of ABE or CBE. h Preference of position flow for targeted As or Cs in the protospacer of target sites for Endo-specific and Inte-specific editing outcomes.