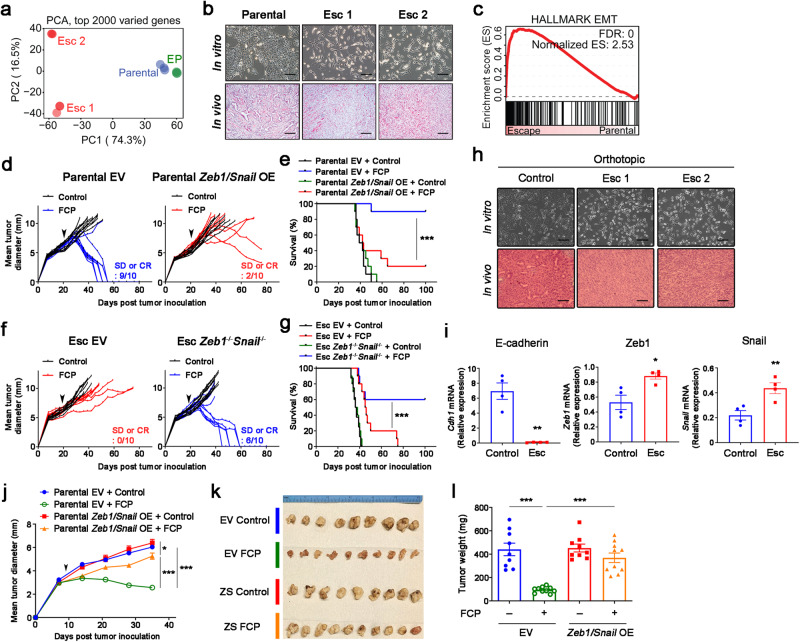
Fig. 2. EMT induces immunotherapy resistance in PDAC.
a PCA plot of RNA-seq data from parental, EP, and Esc cell lines (triplicates). b Representative bright field (top) and H&E (bottom) images of cultured cells and s.c. implanted tumors on day 18. Scale bars, 250 µm. c GSEA of the EMT Hallmark (Molecular Signature Database) in 4662 parental vs. Esc cell lines. Normalized enrichment score (NES) and false discovery rate (FDR) are shown. Individual tumor growth (d) and survival (e) of mice bearing s.c. implanted 4662 parental empty vector (EV, left) and Zeb1/Snail OE (right) tumors treated with either control IgG or FCP (arrow) (n = 10). Individual tumor growth (f) and survival (g) of mice bearing s.c. implanted 4662 Esc EV (left) and Zeb1-/-Snail-/- (right) tumors treated with either control IgG or FCP (arrow) (n = 10). SD, stable disease. Clonal 4662 (C7 and C10) and derived Esc (C7.e1 and C10.e1) lines were used for genetic modification, and both lines showed a similar phenotype. h Representative bright field (top) and H&E (bottom) images of in vitro and in vivo (orthotopic) control and Esc tumors following treatment with control IgG and immunotherapy (FCP), respectively. Scale bars, 250 µm. i qPCR of E-cad, Zeb1, and Snail in control and Esc tumor cell lines established from orthotopic tumors (n = 4). Mean tumor diameter over time by ultrasound imaging of orthotopic 4662 EV or Zeb1/Snail OE tumors treated with control IgG or FCP (arrow) (j). Bright field dissection images (k) and tumor weights (l) of orthotopic pancreas tumors 6 weeks post tumor implantation (n = 9 for controls and 10 for FCP). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.0001 by log-rank (Mantel-Cox) test (e, g), Student’s t test (i), and one-way ANOVA (j, l). Data represent two independent experiments. Source data and exact P value are provided as a Source Data file.