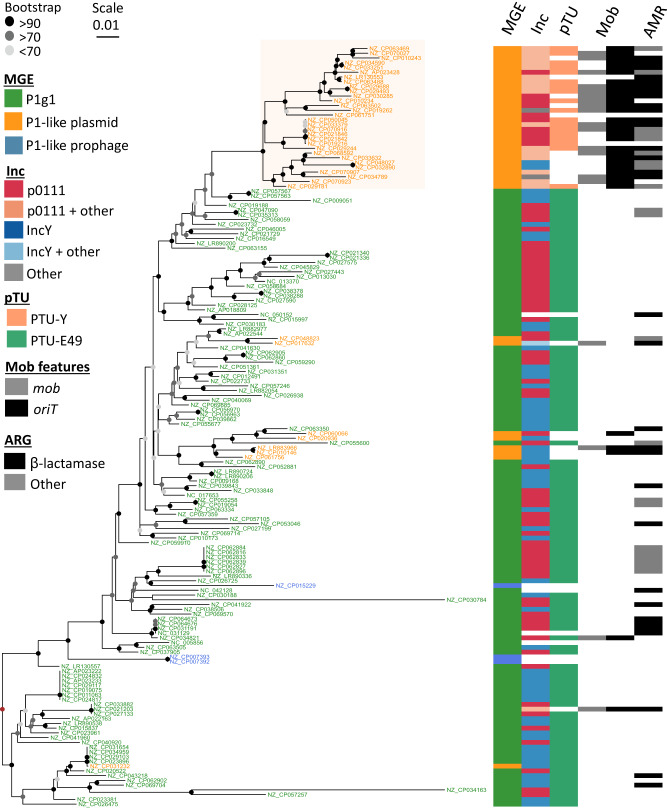
Fig. 6. Phylogenetic tree of P1-like mobile elements.
A global alignment of the 17 persistent gene families was done and given to IQ-TREE v2 (see “Methods”) to compute a phylogenetic tree by maximum likelihood with 1000 ultrafast bootstraps (best model: SYM + I + G4). Visualization was done with ggtree89 in the R environment. Origins of transfers and relaxases were searched in plasmids and phage-plasmids in a previous work48. The tree was rooted using the method of the midpoint-root (brown node) and confidence of the bootstrap values are indicated by the node colors ranging from light gray (less than 70%, uncertain branches) over gray (>70–90%, moderate) to black (>90%, confident branches). The NCBI accessions IDs of the elements are indicated for plasmids and P-Ps in the terminal branches of the tree. For integrative prophages, the ID of the bacterial chromosome is shown with details on the prophage given in Supplementary Dataset S7.