Abstract
The sporadic occurrence of human infections with swine-origin influenza A(H3N2) viruses and the continual emergence of novel A(H3N2) viruses in swine herds underscore the necessity for ongoing assessment of the pandemic risk posed by these viruses. Here, we selected three recent novel swine-origin A(H3N2) viruses isolated between 2017 to 2020, bearing HAs from the 1990.1, 2010.1 or 2010.2 clades, and evaluated their ability to cause disease and transmit in a ferret model. We conclude that despite considerable genetic variances, all three contemporary swine-origin A(H3N2) viruses displayed a capacity for robust replication in the ferret respiratory tract and were also capable of limited airborne transmission. These findings highlight the continued public health risk of swine-origin A(H3N2) strains, especially in human populations with low cross-reactive immunity.
Brief summary:
We used the ferret model to assess the public health risk posed by three swine-origin influenza A(H3N2) viruses isolated in the U.S from 2017–2020, highlighting the necessity of ongoing swine influenza virus surveillance and risk assessment efforts for pandemic preparedness.
Since 2010, there have been 439 confirmed cases of human infection with swine-origin influenza A(H3N2) variant (v) viruses in the United States, one of which has resulted in death [1]. During this time, A(H3N2)v viruses have exhibited considerable genetic and antigenic variances, necessitating regular updates of risk assessment evaluations and pre-pandemic candidate vaccine strain selection. Currently, multiple genotypes of influenza A viruses (IAV) of the H3N2 subtype are cocirculating in U.S. swine populations; the dominant circulating viruses include those belonging to the 1990.4, 2010.1 and 2010.2 H3 HA swine clades and 1998 and 2002 N2 NA swine clades (Global H3 HA and N2 NA swine influenza virus nomenclature identifies a clade based on the decade, or year for HA or NA, when its ancestral human seasonal A(H3N2) virus or swine live attenuated vaccine strain first spilled over to swine and established sustained transmission in pigs) [2, 3]. Since their introduction, the major HA and NA clades of swine A(H3N2) virus have further evolved in swine hosts, splitting into statistically supported monophyletic clades; e.g., N2.2002A and 2002B [4].
In 2017, a swine A(H3N2) virus isolate bearing an HA and NA derived from the circulating human seasonal A(H3N2) virus (from the 2016–2017 Northern Hemisphere influenza season) and internal genes from endemic swine virus was first detected in diagnostic samples from pigs submitted from Oklahoma; this reassortant virus was later found to be endemic in U.S. swine [3, 5]. The HA of this genotype was designated as the H3.2010.2 clade to mark the second human A(H3N2) virus established in swine during the 2010 decade [6]. Before 2017, the majority of recorded A(H3N2)v viruses in the U.S. possessed an HA from the 1990.4 or 2010.1 clades paired with a 2002 or 2002A N2 NA clade [7–9]; risk assessment studies with a focus on virus pathogenicity and transmission in a ferret model suggested that these viruses pose a moderate risk to humans [8, 9]. However, in recent years, swine A(H3N2) viruses bearing a 2010.1 HA paired with 2002B N2 NA and swine internal genes have gained increased dominance in U.S. swine, leading to documented zoonotic transmission events at agricultural fairs or exhibitions in five states in 2017 [10]. Beyond viruses with 2010.1 and 2010.2 HA genes, a novel clade 1990.1 A(H3N2)v reassortant possessing 6 gene segments (PB1, PA, HA, NA, NP, M) from the live attenuated vaccine strain, A/swine/Texas/4199–2/1998 (Tx98), and two segments (NS, PB2) from the A(H1N1) 2009 pandemic virus (H1N1pdm09) was associated with a human infection in Hawaii in 2020, highlighting the continued diversity of A(H3N2)v viruses capable of causing disease in humans in various geographic locations.
To assess the pandemic risk posed by these newly emerged swine A(H3N2) viruses, we studied three contemporary swine-origin A(H3N2) viruses, encapsulating genotypes associated with recent human infections or increased proportionality in US swine. The clade 2010.1 A(H3N2)v virus, A/Ohio/13/2017 (OH/17, GSAID isolate ID # EPI_ISL_277234), was isolated from a human infection case associated with prior exposure to swine at an agricultural fair in Ohio in 2017. The clade 1990.1 A(H3N2)v virus, A/Hawaii/28/2020 (HI/20, GISAID isolate ID# EPI_ISL_501166) was isolated from a patient with no prior known exposure to swine. A swine-origin A(H3N2) virus, A/swine/Oklahoma/A02218157/2017 (SW/OK/17, GISAID isolate ID # EPI_ISL_278441), was isolated from a swine specimen submitted to the Iowa State University Veterinary Diagnostic Laboratory in 2017 and was included to represent the newly established H3.2010.2 lineage. The HA and NA pairing was unique for each virus, and their HA and NA clades and internal gene constellations are listed in Table 1. All viruses were characterized for their ability to cause disease and transmit in a ferret model, which is considered the gold standard small mammalian model for concurrent assessments of IAV pathogenicity and transmission in humans.
Table 1.
Genetic characteristics of swine-origin A(H3N2) viruses.
| Virus | Name in the study | Sub-type | HA genetic clade | NA genetic clade | M gene lineage | amino acid residues | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| HA antigenic sitesa | HA receptor binding site (190/226/228) | PB2 (590/ 591) |
PB2 (627/ 701) |
|||||||
| A/Ohio/13/2017 | OH/17 | H3N2v | 2010.1 | 2002B | H1pdm09 | NTHNFK | D/I/S | S/R | E/D | |
| A/Swine/Oklahoma/A02218157/2017 | SW/OK/17 | H3N2 | 2010.2 | 2016 human seasonal H3N2 virus like | H1pdm09 | STHNYK | D/I/S | S/R | E/D | |
| A/Hawaii/28/2020 | HI/20 | H3N2v | 1990.1 | 1998 | A/swine/Texas/98 LAIV like | KHKHNY | E/I/S | S/R | E/D | |
HA antigenic site refers to residues at position 145, 155, 156, 158, 159, 183.
To evaluate the ability of each of the three viruses to cause disease and spread in humans in a ferret model, 9 male ferrets (6 to 9 months old) per group were intranasally inoculated with 6.0 log10 PFU of virus, among which 6 ferrets were monitored daily for weight loss, signs of disease, and virus shedding in nasal wash specimens as previously described [9]. These animals served as donors for transmission assessments, with a naïve ferret housed either in the same cage (direct contact transmission model, n=3) or in an adjacent cage with perforated side walls (respiratory droplet transmission model, n=3) at a 1:1 donor: contact ratio for each inoculated animal [9]. The remaining three virus-inoculated ferrets per virus group were euthanized on day 3 post inoculation (p.i.) to evaluate virus replication in the respiratory tract and the ability to spread systemically. Among the three viruses evaluated, OH/17 virus caused the greatest weight loss (mean maximum weight loss of 14.3%), whereas SW/OK/2017 and HI/20 virus infection only led to marginal weight loss (mean weight loss of approximately 5–6%) (Table 2). All inoculated animals exhibited only mild symptoms including a slight temperature rise (ranging from 1.1 to 1.6 °C above baseline) and intermittent sneezing or nasal discharge. All virus-inoculated ferrets, regardless of the inoculation strain, shed high titers of infectious virus (mean maximum viral titer of approximately 6.0 log10 PFU/ml) in nasal wash samples between days 1–3 p.i., with the exception of one SW/OK/17 virus-inoculated ferret that exhibited a delay in viral replication before reaching comparable mean peak titers as other animals within the group (Fig 1 & Table 2). Consistent with robust nasal virus shedding in nasal washes, all three viruses replicated efficiently in the ferret upper respiratory tract day on 3 p.i., as supported by mean viral titers ranging from 3.4–5.4 log10 PFU/ml in nasal turbinate tissue (Fig 2). In contrast, viral replication in the lower respiratory tract (including trachea and lung tissues) was detected at reduced frequency and magnitude; OH/17 virus was detected only in the trachea in 2/3 ferrets (mean viral titer 4.6 log10 PFU/g), SW/17 virus was detected only in the lung in 2/3 ferrets (mean viral titer 4.6 log10 PFU/g), and HI/20 virus replicated in both lung and trachea in 1/3 ferrets at viral titers of 3.2–3.5 log10 PFU/g (Fig 2). Sporadic detection of infectious virus at low titers (ranging from 2.2 to 3.5 log10 PFU/g) was observed in olfactory bulb and brain tissues, with no viral replication detected in other extra pulmonary tissues (kidney, spleen, liver, intestine) (Fig 2).
Table 2.
Clinical signs, replication, and transmission of swine-origin A(H3N2) viruses in ferrets
| Virus | Ferret groupsa | Mean max weight loss (range of days)b | Mean max increase in temp (range of days)c | No. of animals with nasal discharge/total No. of animals | No. of animals with sneezing/total No. of animals | Mean max viral titer in NW (log10 PFU/ml)d | No. of sero-conversion/total No. of animalse |
|---|---|---|---|---|---|---|---|
| OH/17 | inoculated | 14.3 (6–12) | 1.6 (1–12) | 3/6 | 4/6 | 6.1 | 6/6 (320–1280) |
| contact (DCT) | 10.7 (9–11) | 1.2 (3–9) | 2/3 | 2/3 | 5.2 | 3/3 (640) | |
| contact (RDT) | 16.1 (9) | 1.6 (6) | 0/3 | 0/3 | 5.4 (1/3) | 2/3 (160–640) | |
| SW/OK/2017 | inoculated | 4.9 (6–12) | 1.3 (1–2) | 0/6 | 6/6 | 6.2 | 6/6 (320–1280) |
| contact (DCT) | 1.1 (8) | 0.9 (2–7) | 1/3 | 2/3 | 6.1 | 3/3 (640–1280) | |
| contact (RDT) | 0.7 (9) | 0.7 (−9) | 0/3 | 0/3 | 6.0 (1/3) | 2/3 (320–640) | |
| HI/20 | inoculated | 5.9 (2–12) | 1.1 (2–12) | 0/6 | 6/6 | 6.3 | 6/6 (320–1280) |
| contact (DCT) | 2.6 (5–9) | 1.1 (4–8) | 1/3 | 2/3 | 5.5 | 3/3 (1280) | |
| contact (RDT) | 5.0 (9) | 0.6 (4) | 0/3 | 0/3 | 5.7 (1/3) | 1/3 (1280) |
Inoculated: animal intranasally inoculated with 6.0 log10 PFU of virus; DCT: direct contact transmission model; RDT: respiratory droplet model.
Percent mean maximum weight loss in inoculated animals (n=6) or contact animals from DCT experiments (n=3 each) or contact animals from RDT experiments with detectable virus shedding (n=1), with the range of days associated with maximum weight loss shown in parentheses.
Mean maximum temperature increase over the baseline from inoculated animal (n=6) or contact animals from DCT experiments (n=3) or contact animals from RDT experiments with detectable virus shedding (n=1), with the range of days associated with maximum temperature rise shown in parentheses.
Mean maximum viral titers in nasal wash specimens from ferrets (log10 PFU/ml). Mean titer is inclusive of all animals per group unless specified in parentheses, where the total number of contact animals with detectable virus in this specimen/total number of animals in the group is stated.
Ferret serum samples were collected on days 21 p.i. or p.c. and tested against the homologous virus in the HI assay. The number of ferrets that seroconverted following inoculation or contact/the total number of ferrets is shown. The limit of detection for the HI assay was 1:10.
Fig 1.
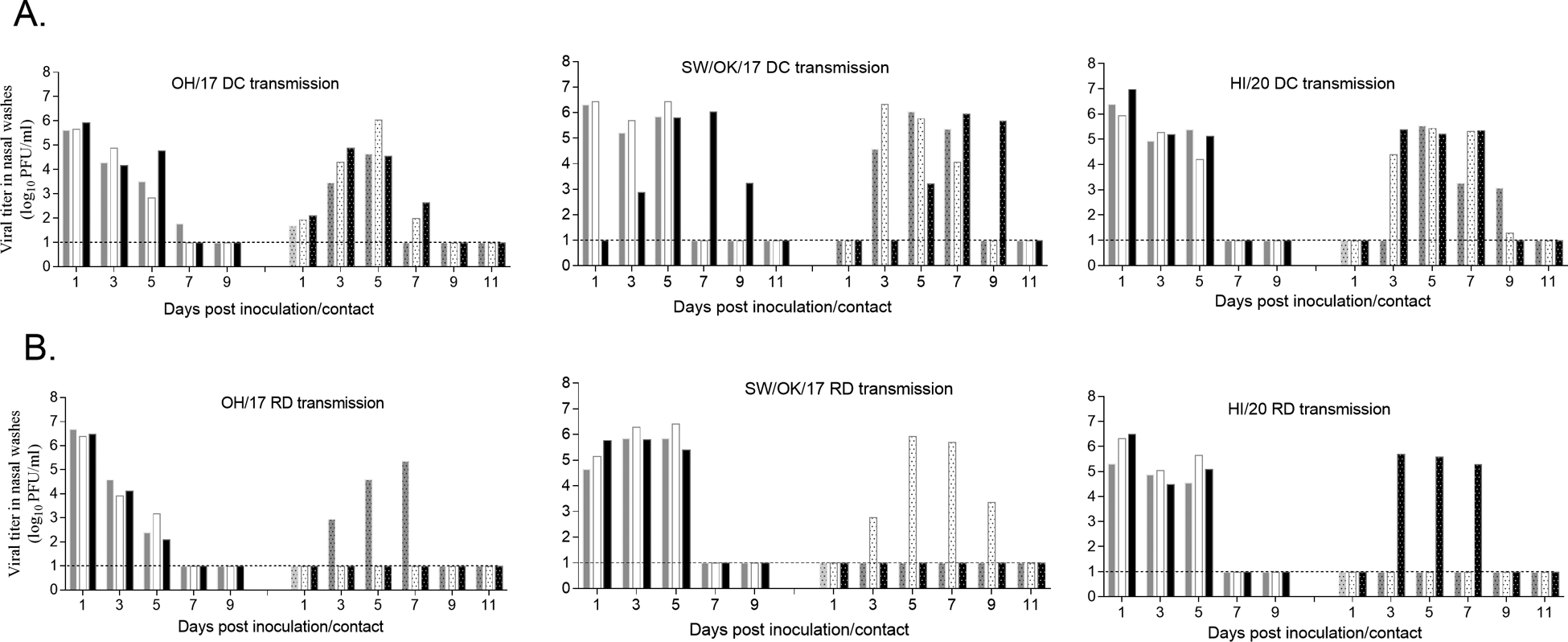
Swine-origin A(H3N2) virus transmission by direct contact (DC) or by respiratory droplets (RD) in the ferret model. Three naïve male ferrets (6–9 months old) were each pair housed with a male ferret (6–9 months old) inoculated with OH/17, SW/OK/17, or HI/20 at 24 hr p.i. in the same cage for evaluating virus transmission via direct contact (A) or placed in an adjacent cage 24 hr p.i. with perforated side walls for evaluating virus transmission by respiratory droplets (B). Nasal wash specimens from inoculated or contact ferrets were collected on alternating days 1–11 p.i. or p.c. and viral titers in the nasal wash specimens were determined in MDCK cells by standard plaque assay and shown in bar graphs with inoculated ferret sample titers shown in left and contact ferret sample titers shown in right panels. Dashed line indicates the limit of detection (1 log10 PFU/ml).
Fig 2.
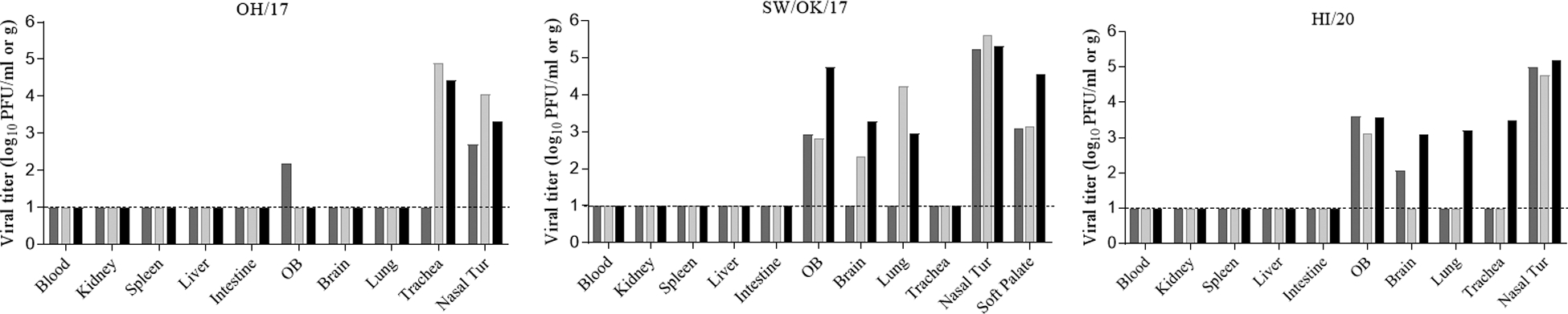
Swine-origin A(H3N2) virus replication in ferret tissues. Three naïve ferrets (6–9 months old) each were intranasally inoculated with 6.0 log10 PFU of viruses and ferret blood or tissues including nasal turbinate (Nasal Tur), trachea, lung, brain (pooled from both anterior and posterior sections), olfactory bulb (OB), kidney, spleen, liver, and intestine (pooled from duodenum, jejunoileal loop, and descending colon) were collected on day 3 p.i. for viral titer determination in MDCK cells by standard plaque assay. Viral titers are expressed at log10 PFU per ml (for nasal turbinate, olfactory bulb) or per gram of tissue. Dashed line indicates the limit of detection (1 log10 PFU/ml or g).
With regard to virus transmission, all three swine-origin A(H3N2) viruses transmitted efficiently via direct contact, with contact ferrets shedding virus with similar peak viral titers and kinetics on day 3–9 post contact (p.c.) as inoculated ferrets; all contact ferrets from these experiments seroconverted to homologous virus (Fig 1 & Table 2). Furthermore, all three viruses exhibited a limited capacity for transmission by respiratory droplets. One of three contact ferrets in each virus group became productively infected, as evidenced by detection of infectious virus in NW specimens on day 3 p.c. with subsequent seroconversion to homologous virus; one additional contact ferret in both OH/17 and SW/OK/17 virus groups also exhibited seroconversion by day 21 p.c. in the absence of infectious virus detection (Fig 1 and Table 2).
Collectively, our risk assessment study concludes that despite genetic divergence, newly emerged swine A(H3N2) viruses detected in the U.S from 2017–2020 are capable of efficient viral replication in the ferret upper respiratory tract, shedding viruses at high titers in nasal wash specimens. These viruses can be efficiently transmitted via direct contact and are also capable of limited transmission by respiratory droplets. Our study provides supportive evidence to the CDC Influenza Virus Risk Assessment Tools (IRAT) for OH/17 virus, determined by this rubric to be of moderate pandemic risk, comparable to the previously assessed A/Indiana/08/2011 virus with a 1990.4 (previously referred to as Cluster IV) lineage HA and 2002A N2 NA [11]. Currently, the 2010.1 HA and 2002B NA pairing represented by OH/17 virus has shown a steadily increasing prevalence in U.S. swine, likely due to the selection pressure posed by swine population immunity and interstate movement of pigs [4]. 2002A and 2002B NA lineages have a greater than 6% genetic distance [4]. A shift in the N2 NA pairing from 2002A to 2002B with 2010.1 H3 HA viruses did not appear to have a substantial effect on virus replicative ability in ferrets, as OH/17 virus caused similar weight loss and virus shedding as the previously characterized H3N2v virus A/Ohio/16 (OH/16) virus; OH/16 virus shares an identical internal gene constellation as OH/17 virus and caused approximately 16.5% mean maximum weight loss in ferrets, reaching6.1 log10 PFU/ml in mean peak viral titer in ferret nasal wash specimens [9], but with 2010.1 HA and 2002A NA pairing. However, OH/16 virus was able to transmit by respiratory droplets with productive virus shedding and seroconversion in 3/3 contact ferrets in the same experimental setting compared to OH/17 virus, which only resulted in productive virus shedding in 1/3 contact ferrets, suggesting that the functional balance of HA and NA is important for airborne transmission of swine IAV in the ferret model. SW/OK/17 virus represents the virus with the HA and NA from the most recent incursion of human seasonal A(H3N2) virus that have established sustained transmission in the U.S. swine population; the virus shares six key residues with the human A(H3N2) vaccine strain from the 2014 season which are critical for virus antigenicity [12]. Similar to human seasonal A(H3N2) virus [13], SW/OK/17 virus infection in ferrets only resulted in marginal weight loss despite efficient virus replication in the upper respiratory tract. However, unlike human seasonal A(H3N2) viruses [13], SW/OK/17 virus does not possess a full capacity for airborne transmission in the ferret model, suggesting that internal genes also play a role in swine influenza virus transmission in mammalian hosts.
HI/20 virus represents a novel reassortant between a swine LAIV strain and the H1N1pdm09. Bivalent swine LAIV vaccine strains consisting of A/swine/Minnesota/37866/1999 (H1N1) and TX98 (H3N2) on the backbone of TX98 virus were first commercialized in the U.S in 2017 and constructed by incorporating a deletion in the nonstructural protein (NS1) for reduced anti-interferon activity, resulting in viruses with significantly reduced replicative capacity in swine hosts [14]. Following the use of LAIV as a vaccine strategy for swine influenza virus control in the U.S., swine A(H3N2) viruses with the HA and NA from TX98-like virus and one or more internal gene segments from endemic swine viruses have been detected in swine specimens [5]. The acquisition of NS and PB2 segments from the H1N1pdm09 virus likely contributes to the efficient viral replication and limited airborne transmission associated with HI/20 virus in the ferret model. Currently, the extent of potential reassortment between LAIV strains and endemic swine viruses and the impact these reassortment events contribute to virus genetic ecology in swine herds remain unclear. The emergence of reassortant A(H3N2)v virus (HI/20), which has the HA and NA gene segments from TX98-like virus and internal genes from endemic swine or H1N1pdm09 virus, coupled with the lack of sufficient cross-reactive antibodies against historic H3 viruses especially in younger age groups [12, 15], highlights the zoonotic risk posed by these types of reassortant viruses.
Taken together, our study confirms that robust viral replication and a limited ability to spread by the airborne route are associated with newly emerged swine A(H3N2) viruses. Although the HA of these viruses is descendant from human seasonal H3 virus, their evolution in swine may take a pathway different from the A(H3N2) viruses in humans. Considering differences in seroprotection rates against historic H3N2 viruses across the human population [12, 15], the public health threat posed by swine influenza viruses cannot be ignored. Continued swine influenza virus surveillance and risk assessment of contemporary IAV at the swine-human interface are essential for pandemic preparedness.
Acknowledgements.
The findings and conclusions are those of the authors and do not necessarily reflect the views of ASTDR/the Centers for Disease Control and Prevention (CDC).
References:
- 1.CDC. Novel Influenza A Virus Infections. Available at: https://gis.cdc.gov/grasp/fluview/Novel_Influenza.html. Accessed April 10 2023.
- 2.Neveau MN, Zeller MA, Kaplan BS, et al. Genetic and Antigenic Characterization of an Expanding H3 Influenza A Virus Clade in U.S. Swine Visualized by Nextstrain. mSphere 2022; 7:e0099421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeller MA, Anderson TK, Walia RW, Vincent AL, Gauger PC. ISU FLUture: a veterinary diagnostic laboratory web-based platform to monitor the temporal genetic patterns of Influenza A virus in swine. BMC Bioinformatics 2018; 19:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeller MA, Chang J, Vincent AL, Gauger PC, Anderson TK. Spatial and temporal coevolution of N2 neuraminidase and H1 and H3 hemagglutinin genes of influenza A virus in US swine. Virus Evol 2021; 7:veab090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma A, Zeller MA, Li G, et al. Detection of live attenuated influenza vaccine virus and evidence of reassortment in the U.S. swine population. J Vet Diagn Invest 2020; 32:301–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma A, Zeller MA, Souza CK, et al. Characterization of a 2016–2017 Human Seasonal H3 Influenza A Virus Spillover Now Endemic to U.S. Swine. mSphere 2022; 7:e0080921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox CM, Neises D, Garten RJ, et al. Swine influenza virus A (H3N2) infection in human, Kansas, USA, 2009. Emerg Infect Dis 2011; 17:1143–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearce MB, Jayaraman A, Pappas C, et al. Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proc Natl Acad Sci U S A 2012; 109:3944–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun X, Pulit-Penaloza JA, Belser JA, et al. Pathogenesis and Transmission of Genetically Diverse Swine-Origin H3N2 Variant Influenza A Viruses from Multiple Lineages Isolated in the United States, 2011–2016. J Virol 2018; 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duwell MM, Blythe D, Radebaugh MW, et al. Influenza A(H3N2) Variant Virus Outbreak at Three Fairs - Maryland, 2017. MMWR Morb Mortal Wkly Rep 2018; 67:1169–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.CDC. Summary of Influenza Risk Assessment Tool (IRAT) Results. Available at: https://www.cdc.gov/flu/pandemic-resources/monitoring/irat-virus-summaries.htm. Accessed April 1 2023.
- 12.Souza CK, Anderson TK, Chang J, et al. Antigenic Distance between North American Swine and Human Seasonal H3N2 Influenza A Viruses as an Indication of Zoonotic Risk to Humans. J Virol 2022; 96:e0137421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maines TR, Chen LM, Matsuoka Y, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A 2006; 103:12121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solorzano A, Webby RJ, Lager KM, Janke BH, Garcia-Sastre A, Richt JA. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J Virol 2005; 79:7535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorbach JN, Fitzgerald T, Nolan C, et al. Gaps in Serologic Immunity against Contemporary Swine-Origin Influenza A Viruses among Healthy Individuals in the United States. Viruses 2021; 13. [DOI] [PMC free article] [PubMed] [Google Scholar]


