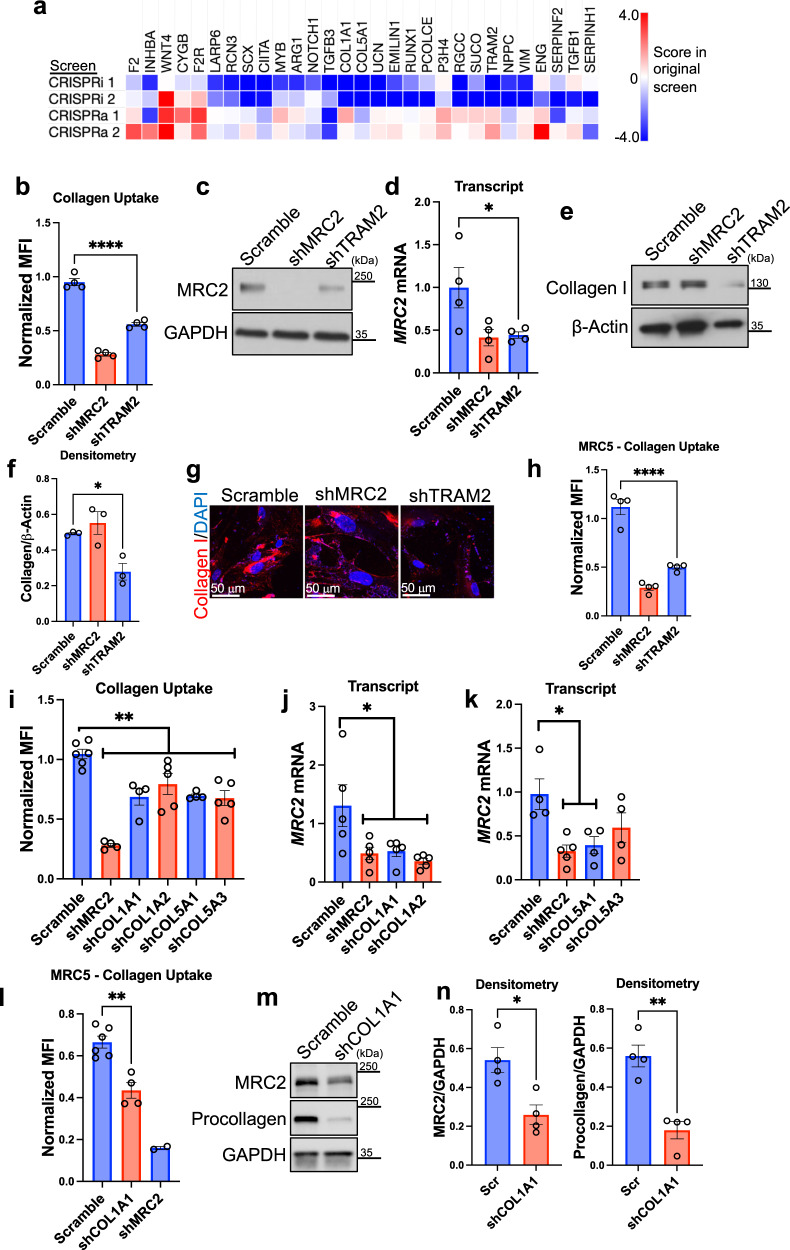
Fig. 3. Collagen biosynthesis positively regulates cell-based collagen clearance and MRC2 expression.
a Heatmap of overall phenotype scores from original screens as indicated for top genes from ‘Collagen Biosynthetic Process’ GO gene-set (displayed genes had most extreme ‘Score’ values in one or more screen result, with most extreme ‘Score’ absolute value at least >1 and concordant with GO term; 29/43 genes from this GO term that were included in the screen fit these criteria and are displayed). b Flow-cytometry based collagen-uptake assay in U937 cells after shRNA treatment as indicated vs. Scramble control. MFI Mean Fluorescence Intensity. N = 4 per group; p < 0.0001. c Western blot of U937 cells after shRNA treatment as indicated vs. Scramble control, representative of N = 4 independent experiments. d Q-RT-PCR in U937 cells after shRNA treatment as indicated vs. Scramble control. N = 4 per group; p = 0.0483. e, f Western blot and densitometry of MRC5 fibroblasts after shRNA treatment as indicated vs. Scramble control, representative of N = 3 independent experiments; p = 0.0477. g Representative confocal immunofluorescence images of MRC5 fibroblasts after shRNA treatment as indicated vs. Scramble control. h Flow-cytometry based collagen-uptake assay in MRC5 cells after shRNA treatment as indicated vs. Scramble control. N = 4 per group; p < 0.0001. i Flow-cytometry based collagen-uptake assay in U937 cells after shRNA treatment as indicated vs. Scramble control. N = 6 (Scramble), 4 (shMRC2, shCOL1A1, shCOL5A1), 5 (shCOL1A2, shCOL5A3) independent biological replicates; p-values are for post-hoc testing of Scramble vs. the following: shMRC2 (p < 0.0001), shCOL1A1 (p = 0.0005), shCOL1A2 (p = 0.0088), shCOL5A1 (p = 0.0006), shCOL5A3 (p = 0.0001). (j–k) Q-RT-PCR in U937 cells after shRNA treatment as indicated vs. Scramble control. N = 5 (left, all groups), 4 (right, Scramble, shCOL5A1, shCOL5A3), 5 (right, shMRC2) independent biological replicates; j p = 0.0136 (Scramble vs. shMRC2), 0.0189 (Scramble vs. shCOL1A1), 0.0041 (Scramble vs. shCOL1A2); k p = 0.0080 (Scramble vs. shMRC2), 0.0223 (Scramble vs. shCOL5A1). l Flow-cytometry based collagen-uptake assay in MRC5 fibroblasts after shRNA treatment as indicated vs. Scramble control. N = 6 and 4 in Scramble and shCOL1A1 groups respectively, N = 2 in shMRC2 group; p = 0.0011. m, n Western blot and densitometry of MRC5 fibroblasts after shRNA treatment as indicated vs. Scramble control, representative of N = 4 independent experiments; p = 0.0137 (left), p = 0.0017 (right). Data are shown as the mean ± SEM. Statistics: b–k one-way ANOVA with post-hoc Dunnett’s testing; (l–n) unpaired Student’s t test (two-sided). *p < 0.05, **p < 0.01, ****p < 0.0001. Source data are provided as a Source Data file.