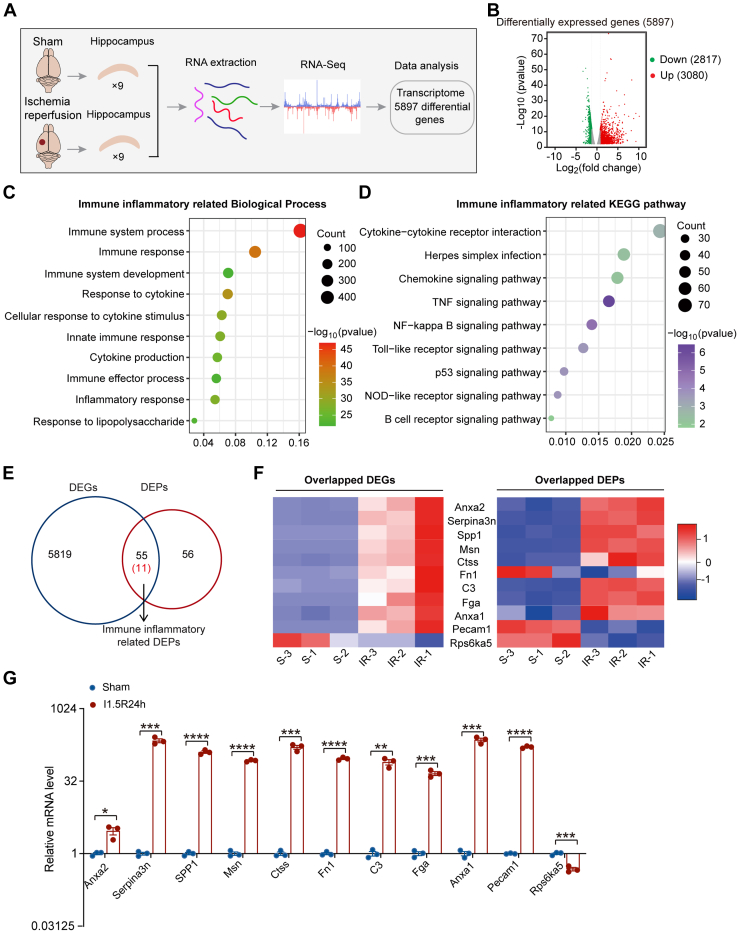
Fig. 2.
Combinatorial analyses of transcriptome, proteome, and phosphoproteome profiles revealed Anxa2 was related to I/R-triggered inflammatory response.A, schematic diagram of transcriptome profile. Three samples of hippocampi for each replicate after sham operation or ischemia 90 min reperfusion 24 h were analyzed by RNA-seq. B, volcano plot of the differential gene expression between sham and I/R groups (fold change >1.2 or <1/1.2, p-value <0.05). C and D, top-ranked biological processes (C) and KEGG pathways (D) related to immune inflammatory responses enriched for differentially expressed genes (DEGs) after I/R in transcriptome profile (p-value <0.05). E, Venn diagram of common DEGs and DEPs associated with the immune inflammatory response between transcriptome and proteome. F, heatmap of the expression trends of common DEGs and DEPs related to immune inflammatory responses between I/R versus sham. Normalized and z-scored (color-coded) value of log2 PFKM of each gene or log2 expression value of each protein is shown. Red and blue colors represent upregulated and downregulated genes, respectively. G, the expression level of immune inflammatory-related genes was analyzed by RT-PCR after I/R relative to Sham. GAPDH was used as the internal control for RT-PCR. Data are presented as means ± SEM, Statistical significance was determined by unpaired student’s t-tests, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Anxa2, annexin A2; DEP, differentially expressed protein; I/R, ischemic reperfusion; KEGG, Kyoto Encyclopedia of Genes and Genomes.