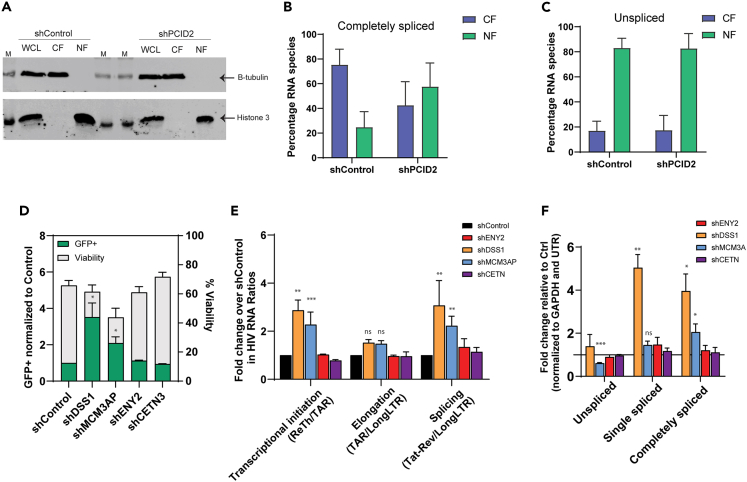
Figure 6.
Effect of PCID2 and other TREX2 complex members on transcriptional and post-transcriptional steps of HIV-1 gene expression during viral latency
(A) Western blot of whole cell lysate (WCL) and cell fractions corresponding to cytoplasm fraction (CF) and nuclear fraction (NF) in shControl or shPCID2 knockdown J-Lat 11.1 to probe for B-tubulin and histone 3. M refers to protein marker.
(B and C) Percentage of RNA species unspliced HIV-1 vRNA (B) and completely spliced HIV-1 vRNA (C) in the cytosolic fraction (CF) and nuclear fraction (NF). Bars and error lines represent mean and SEM (n = 3).
(D) Fold increase in the % of GFP (left y axes) and viability (right y axes) in shControl or shDSS1, shMCM3AP, shENY2, and shCETN3 J-Lat 11.1 cells as measured by flow cytometry. Bars represent mean of three independent shRNA-mediated knockdown experiments and error lines represent SEM. Statistical significance was determined by t test; ∗p < 0.05. Raw values for percentage of GFP are available in Table S2.
(E) Transcriptional profiling assay of shControl and shDSS1, shMCM3AP, shENY2, and shCETN3 J-Lat 11.1 cells. Gene expression blocks at transcriptional initiation, elongation, and post-transcriptional steps are assessed by calculating the ratio of the relative abundance of HIV-1 RNA species as shown in the figure. Data are presented as fold change in HIV-1 RNA ratios in knocked down cells relative to shControl. Bars represent mean of three to five independent shRNA-mediated knockdown experiments and error lines represent SEM. Statistical significance was determined by ANOVA test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(F) Fold change in HIV-1 RNA splicing variants upon DSS1, MCM3AP, ENY2, and CETN3 knockdown relative to shControl. Values were normalized to GAPDH and UTR HIV-1 transcript. Bars represent mean and error lines represent SEM. Statistical significance was determined by t test; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.