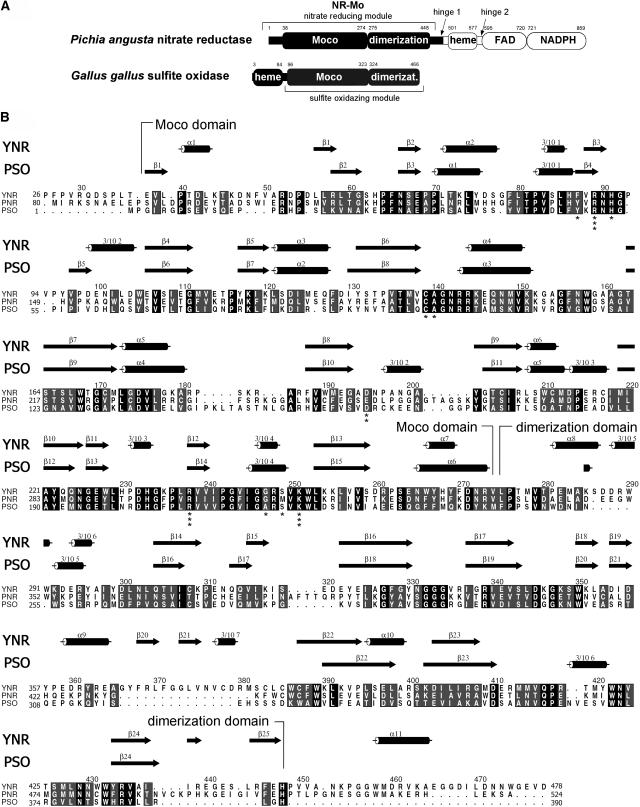
Figure 1.
Primary and Secondary Structure Comparison of NR and SO.
(A) Domain structure of P. angusta NR and CSO. The NR-Mo fragment that has been expressed, purified, and crystallized is shown in black. The first and last residues of conserved domains (Moco binding, dimerization, heme, FAD, and NADPH binding domains) are indicated.
(B) Sequence alignment of yeast NR (P. angusta, YNR) and Arabidopsis NR2 (plant NR, PNR) with Arabidopsis SO (plant SO, PSO). Strictly conserved residues are highlighted in black, and conserved residues are highlighted in gray. Secondary structure elements for YNR and PSO are shown above the alignment with cylinders for α- or 310-helices and arrows for β-sheets. The alignment was generated with ClustalW (Thompson et al., 1994) and ALSCRIPT (Barton, 1993). Secondary structure elements were determined with PROMOTIF (Hutchinson and Thornton, 1996). Residue numbering is based on the primary sequence of YNR. Residues of NR-Mo that coordinate the Moco are indicated with asterisks. The number of asterisks correlates with the number of contacts to Moco.