Figure 1. Genetic and immunological determinants of COVID-19 pneumonia.
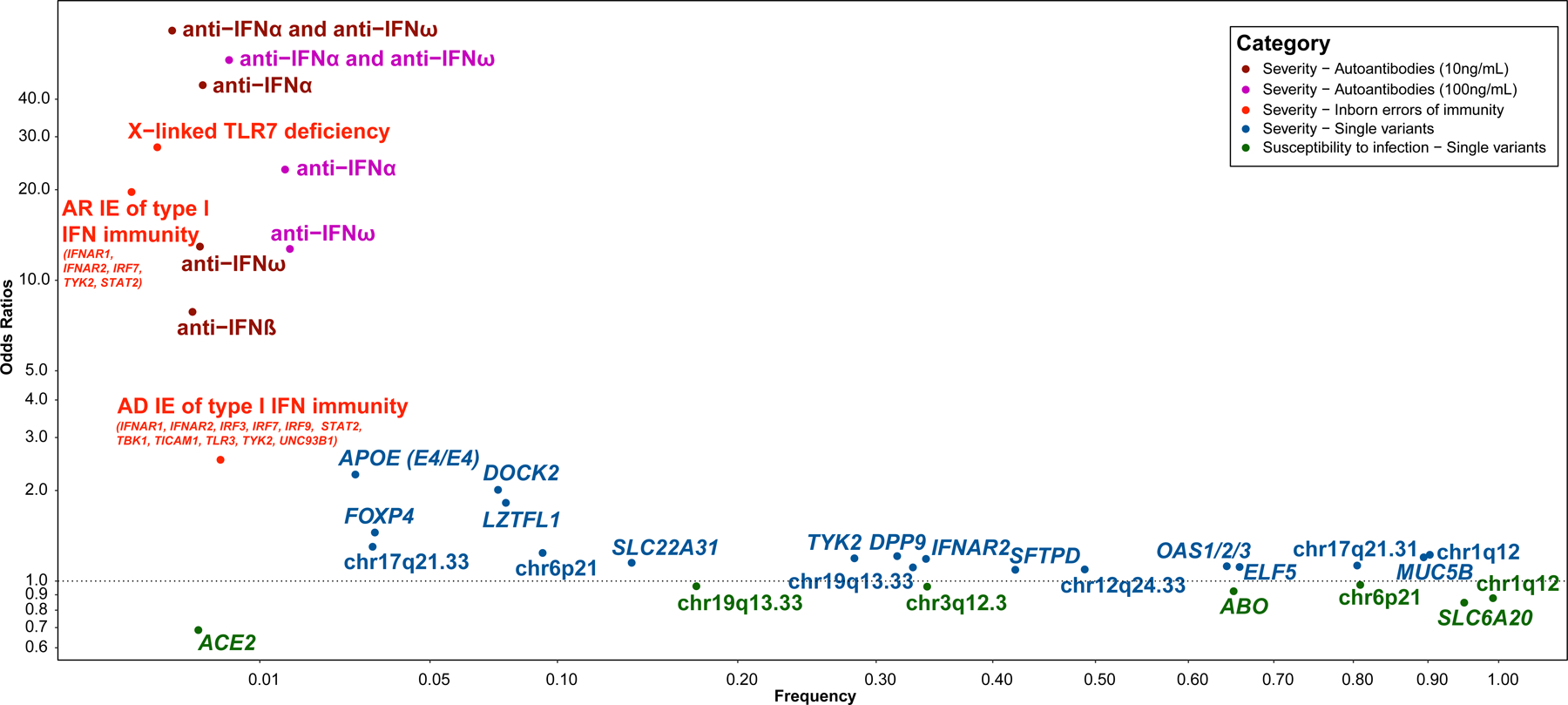
Odds ratios (OR) for the associations of auto-Abs against type I IFN, inborn errors of immunity (IEIs) and single genetic variants with the severity of COVID-19 pneumonia and resistance to SARS-CoV-2 infection are plotted according to risk factor frequency. For auto-Abs against type I IFN, the ORs and frequency were taken from (114). For X-linked TLR7 deficiency, the OR for the aggregated effect of biochemically proven loss of function (LOF) was taken from (74) and the cumulative frequency of biochemically proven LOF was taken from (76). For other IEIs, the OR for the aggregated effect of homozygous (autosomal recessive, AR) or heterozygous (autosomal dominant, AD) predicted LOF was taken from (74) and the corresponding cumulative frequencies were estimated from Gnomad v2.1.1. For single variants, the OR, assuming an additive model, and deleterious allele frequencies were taken from the most recent update of the COVID-19 Host Genetics Initiative GWAS study (43), except for the DOCK2 and APOE loci. The chromosomal region or closest gene is indicated. For the DOCK2 locus, the OR for the effect of the rs60200309-A variant on the severity of COVID-19 pneumonia under an additive model and allele frequency for rs60200309-A were taken from (156). For the APOE locus, the hazard ratio for the effect of APOE4 homozygosity as opposed to APOE3 homozygosity for COVID-19 mortality and the frequency of APOE4 homozygosity were taken from (157).
