Fig. 5 |. Single-cell Hi-C-based techniques measure the variability of enhancer–promoter contacts.
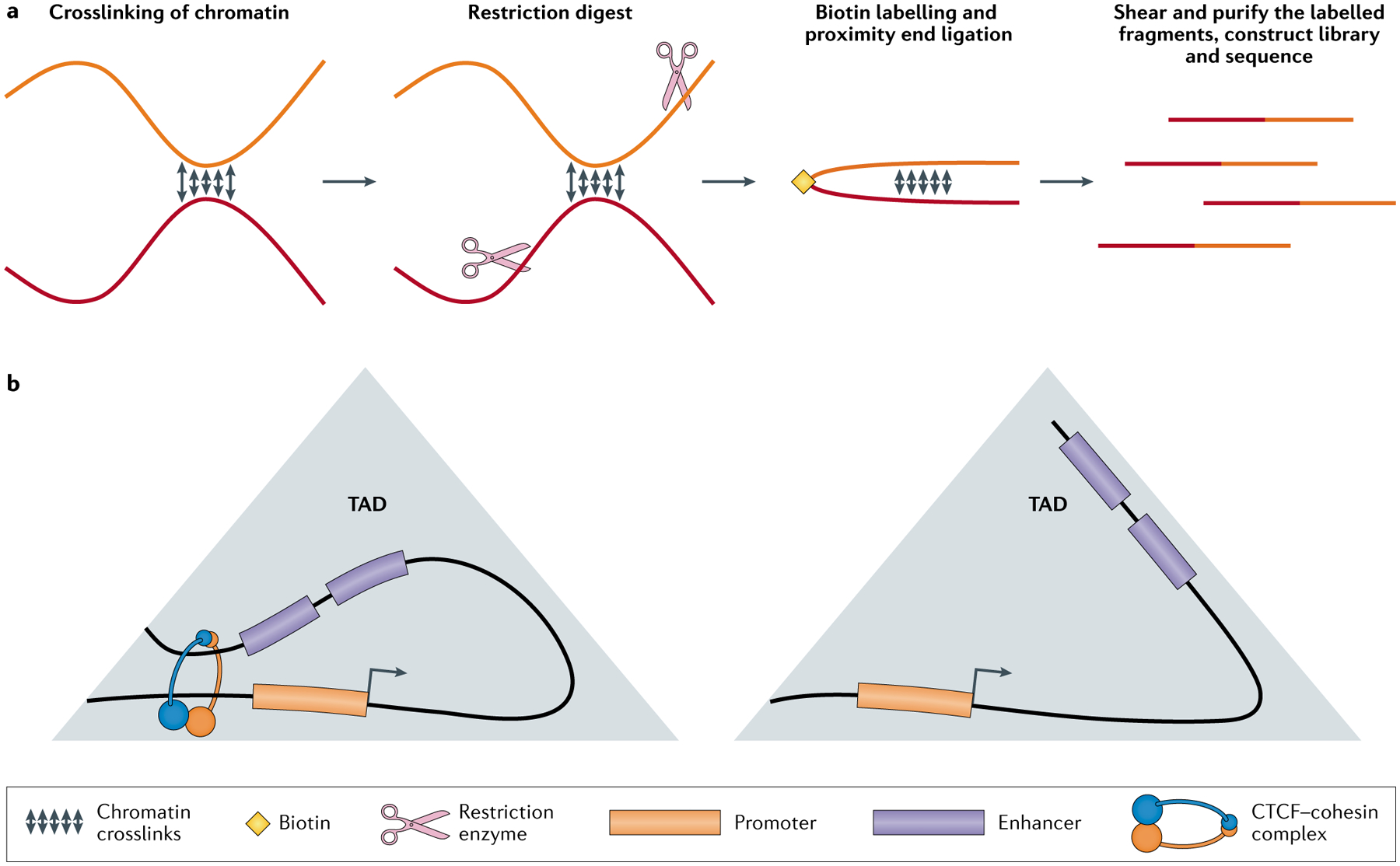
a | Simple depiction of how a chromatin contact is captured by Hi-C. Distal genomic regions in close spatial proximity are indicated using different colours. The chromatin is crosslinked to preserve the interactions, followed by a sequence-specific restriction digest to cut the chromatin into fragments. The cut ends are then ligated together and labelled with biotin. Finally, the crosslinking is reversed and the DNA fragments are purified using the biotin label and sequenced. b | Depiction of topologically associating domains (TADs), which are chromatin regions comprising many intra-regional spatial interactions. TAD exhibiting enhancer–promoter contacts facilitated by CCCTC-binding factor (CTCF) and cohesin. CTCF and cohesin bring distal genomic elements together into domains containing elevated local concentrations of enhancer (orange) and promoter (blue) elements (left). Absence of CTCF-mediated and cohesin-mediated interactions affects transcriptional heterogeneity without disrupting overall TAD structure (right). Hi-C, high-throughput chromosome conformation capture.
