ABSTRACT
Bifidobacteria are among the first microbial colonizers of the human gut, being frequently associated with human health-promoting activities. In the current study, an in silico methodology based on an ecological and phylogenomic-driven approach allowed the selection of a Bifidobacterium adolescentis prototype strain, i.e., B. adolescentis PRL2023, which best represents the overall genetic content and functional features of the B. adolescentis taxon. Such features were confirmed by in vitro experiments aimed at evaluating the ability of this strain to survive in the gastrointestinal tract of the host and its ability to interact with human intestinal cells and other microbial gut commensals. In this context, co-cultivation of B. adolescentis PRL2023 and several gut commensals revealed various microbe-microbe interactions and indicated co-metabolism of particular plant-derived glycans, such as xylan.
IMPORTANCE
The use of appropriate bacterial strains in experimental research becomes imperative in order to investigate bacterial behavior while mimicking the natural environment. In the current study, through in silico and in vitro methodologies, we were able to identify the most representative strain of the Bifidobacterium adolescentis species. The ability of this strain, B. adolescentis PRL2023, to cope with the environmental challenges imposed by the gastrointestinal tract, together with its ability to switch its carbohydrate metabolism to compete with other gut microorganisms, makes it an ideal choice as a B. adolescentis prototype and a member of the healthy microbiota of adults. This strain possesses a genetic blueprint appropriate for its exploitation as a candidate for next-generation probiotics.
KEYWORDS: bifidobacteria, microbiome, genomics, metagenomics, probiotic
INTRODUCTION
The human gastrointestinal tract (GIT) encompasses an intricate community of different microorganisms representing the human gut microbiota (1). Millions of years of co-evolution between the intestinal bacterial community and its host are believed to have contributed to the establishment of multiple trophic interactions, including mutualistic relationships in which the host provides nutrients and a suitable environment for the growth of its intestinal microbes while, in exchange, the latter perform multiple beneficial physiological and metabolic functions (2, 3). Among the different bacterial species residing in the human GIT are members of the genus Bifidobacterium, which represent an extensively studied microbial component due to their purported ability to exert health-promoting or probiotic effects upon their host (4–10).
Bifidobacterium species are considered to be among the first microbial colonizers of the infant gut (11–15), and, despite a decline in their relative abundance from infancy to adulthood, they reach stably maintained numbers until old age, thus in principle capable of eliciting beneficial activities during the entire lifespan of the host (1). Among the reported healthy features exerted by bifidobacteria are maturation of the host immune system (1, 16, 17) and development of intestinal barrier integrity, which assist in protection against pathogen invasion and subsequent proliferation while also maintaining gut homeostasis (15, 18). Furthermore, members belonging to the Bifidobacterium genus have been shown to generate various metabolites, such as short chain fatty acid acetate, polyphenols, vitamins, and conjugated linoleic acids (17, 19). In addition, bifidobacteria can degrade complex host- and diet-derived glycans (20), which facilitates their gut colonization, while it also generates nutrients to both the host and other intestinal commensals through cross-feeding mechanisms (21–23).
Recently, it has been shown that “omics” technologies are crucial when investigating the composition and activities of commensal bacteria (19). In particular, genome analysis of bifidobacterial species instigated a new discipline called probiogenomics (24), which, together with functional genomic data, allows an improved understanding of diversity, evolution, and beneficial effects of commensal bacteria (21, 25, 26). Nonetheless, to date, most bacterial strains included in supplements and added to certain fermented foods like yogurt have been selected based on their superior growth yields and/or survival levels in bioreactor scale production without providing any information on the interplay between bacterial strains and host or with other commensals of the human GIT. Recently, an ecological and phylogenomic-driven approach facilitated the identification of an optimal representative strain or prototype of the Bifidobacterium longum subsp. longum species, i.e., B. longum subsp. longum PRL2022 (19, 27).
Here, we describe a comprehensive screening aimed at identifying a Bifidobacterium adolescentis prototype, i.e., B. adolescentis PRL2023, of the human gut based on the microbiota of 4,019 healthy subjects. The selected prototypical strain was assessed through metatranscriptomic experiments during co-cultivation with bacterial species that co-occur in the human microbiome (28), highlighting how PRL2023 can interact with other commensals inhabiting the intestinal environment (29).
RESULTS AND DISCUSSION
Identification of key bifidobacterial species associated with the human gut of healthy adults
In silico analyses involving 4,019 publicly available human gut microbiome data sets (produced through shotgun metagenomic sequencing) belonging to healthy adults (ranging from 18 to 80 years of age) were investigated to identify the composition of their bifidobacterial communities (Table S1). Microbial profiling based on short-read taxonomic classification down to species level revealed the occurrence of B. adolescentis and B. longum, with a relative abundance of 1.5% ± 0.1% and 1.1% ± 0.1%, respectively, followed by Bifidobacterium pseudocatenulatum (relative abundance of 0.51% ± 0.03%) (Table 1). Among the analyzed human gut metagenomes, the prevalence of the latter species was 42%, 48%, and 25%, respectively, indicating that B. adolescentis and B. longum represent the most common bifidobacterial species across the adult human gut (Table 1). Notably, these same two species were also identified as the most abundant bifidobacterial taxa of the human gut microbiota of a smaller metagenomic data set composed of 76 elderly (healthy humans of 80 years and over) (Table 1). Thus, these findings highlight that B. adolescentis and B. longum are important contributors of the human gut microbiota due to their prevalence and abundance in hosts ranging from adolescence through old age.
TABLE 1.
Abundance of bifidobacteria in healthy subjects
| Adults (>18 years) | Elderly (>80 years) | |||
|---|---|---|---|---|
| Species | Average abundance (%) | Prevalence (%) | Average abundance (%) | Prevalence (%) |
| Bifidobacterium adolescentis | 1.52 ± 0.06 | 42.0 | 1.64 ± 0.23 | 29.5 |
| Bifidobacterium angulatum | 0.06 ± 0.01 | 3.5 | 0.00 ± 0.00 | 0.0 |
| Bifidobacterium animalis | 0.03 ± 0.00 | 2.9 | 0.02 ± 0.01 | 2.7 |
| Bifidobacterium bifidum | 0.26 ± 0.02 | 15.7 | 0.36 ± 0.07 | 15.5 |
| Bifidobacterium breve | 0.02 ± 0.01 | 2.4 | 0.08 ± 0.03 | 6.2 |
| Bifidobacterium catenulatum | 0.13 ± 0.01 | 10.0 | 0.15 ± 0.05 | 6.9 |
| Bifidobacterium dentium | 0.02 ± 0.00 | 2.7 | 0.09 ± 0.02 | 11.8 |
| Bifidobacterium gallinarum | 0.00 ± 0.00 | 0.2 | 0.01 ± 0.00 | 1.1 |
| Bifidobacterium longum | 1.08 ± 0.06 | 47.9 | 1.73 ± 0.23 | 42.2 |
| Bifidobacterium merycicum | 0.00 ± 0.00 | 0.1 | 0.00 ± 0.00 | 0.0 |
| Bifidobacterium pseudocatenulatum | 0.51 ± 0.03 | 25.1 | 0.51 ± 0.13 | 15.2 |
| Bifidobacterium pseudolongum | 0.00 ± 0.00 | 0.2 | 0.00 ± 0.00 | 0.0 |
| Bifidobacterium pullorum | 0.00 ± 0.00 | 0.0 | 0.01 ± 0.01 | 0.4 |
| Bifidobacterium ruminantium | 0.00 ± 0.00 | 0.6 | 0.00 ± 0.00 | 0.2 |
| Bifidobacterium saeculare | 0.00 ± 0.00 | 0.1 | 0.00 ± 0.00 | 0.4 |
| Bifidobacterium scardovii | 0.00 ± 0.00 | 0.2 | 0.00 ± 0.00 | 0.2 |
In silico analyses of the human gut microbiota representing 82 independent metagenomic studies confirmed that B. adolescentis is a typical bifidobacterial colonizer of the adult human gut, as previously shown (30). Furthermore, the dissection of microbial profiles suggests that B. adolescentis and B. longum are the most prevalent and abundant bifidobacterial species inhabiting the gut microbiota of humans across their lifespans. Thus, we focused our interest on identifying a B. adolescentis prototype following a recently published approach (19, 27).
Ecological and phylogenomic-driven identification of B. adolescentis prototypes
To evaluate the distribution of B. adolescentis species among the human gut microbiota, InStrain-based profiling of 113 strains was performed, representing all currently available genome sequences for this bifidobacterial species. First, a de-replication procedure was applied using the dRep software among collected genome sequences, resulting in the identification of 79 distinct genetic lineages of B. adolescentis (Table 2). The occurrence of such a high number of genetically unique strains suggests a high level of genetic heterogeneity within the species, a finding that validates a similar report (31). Thus, their distribution across the gut microbiome of healthy individuals was investigated through a k-mer-based analysis, employing the same above-described data sets used for bifidobacterial profiling to explore. This analysis unveiled the ecological dissemination of each lineage among the metagenomic data sets of healthy adults. When detected, the B. adolescentis strain distribution among microbiomes ranged from 27.9% to 2.9% for PRL2023 and DSM 20087, respectively (Table 2). The wide distribution range of these 79 B. adolescentis lineages highlighted those that commonly inhabit the human gut, and others were rarely identified among the considered population (Table 2).
TABLE 2.
Bifidobacterium adolescentis strain distribution among 4,019 publicly available data sets of the human gut microbiome
| NCBI code | Strain | Average nucleotide identity (between dereplicated genomes) | Prevalence (when detected) (%) | A × P (Average × Prevalence index) |
|---|---|---|---|---|
| GCA_002108035.1 | PRL2023 | 97.88 | 27.9 | 98.77 |
| GCA_003030905.1 | 1–11 | 97.94 | 24.3 | 85.82 |
| GCA_003436185.1 | TM06-4 | 98.07 | 23.5 | 83.33 |
| GCA_019131675.1 | MSK.11.28 | 98.25 | 21.3 | 75.66 |
| GCA_003437735.1 | TF06-2AC | 97.97 | 21.3 | 75.44 |
| Local | 780B | 97.75 | 20.6 | 72.68 |
| GCA_016069975.1 | VKPM Ac-1245 | 97.96 | 19.9 | 70.23 |
| Local | 77B | 98.25 | 18.4 | 65.22 |
| GCA_003466335.1 | TM06-51 | 98.12 | 18.4 | 65.14 |
| GCA_002107975.1 | AL12-4 | 98.01 | 18.4 | 65.06 |
| GCA_019041975.1 | MSK.7.22 | 97.88 | 18.4 | 64.98 |
| GCA_019127835.1 | MSK.20.45 | 98.32 | 17.6 | 62.66 |
| GCA_003472245.1 | AM12-59 | 98.24 | 17.6 | 62.61 |
| GCA_003468385.1 | AM36-3AC | 98.19 | 17.6 | 62.57 |
| GCA_000154085.1 | L2-32 | 98.17 | 17.6 | 62.56 |
| GCA_024460485.1 | SL.1.01 | 98.16 | 17.6 | 62.56 |
| GCA_015553925.1 | BSD2780061687_150420_A4 | 98.23 | 16.2 | 57.38 |
| GCA_002108095.1 | AL46-7 | 98.21 | 15.4 | 54.76 |
| GCA_019972965.1 | 4–2 | 98.19 | 15.4 | 54.75 |
| GCA_000010425.1 | ATCC 15703 | 97.94 | 15.4 | 54.61 |
| GCA_015552825.1 | D53t1_180928_D4 | 98.33 | 14.7 | 52.22 |
| GCA_001406215.1 | 2789STDY5834850 | 98.32 | 14.7 | 52.21 |
| GCA_002107995.1 | LMG 10734 | 98.30 | 14.7 | 52.20 |
| GCA_017815835.1 | PRL2019 | 98.26 | 14.7 | 52.18 |
| GCA_003457765.1 | AF28-4AC | 98.24 | 14.7 | 52.17 |
| GCA_019734235.1 | K09 | 98.23 | 14.7 | 52.17 |
| GCA_018785705.1 | MCC258 | 98.23 | 14.7 | 52.17 |
| GCA_003469145.1 | AM34-11 | 98.13 | 14.7 | 52.11 |
| GCA_015558415.1 | D52t1_170925_B8 | 98.10 | 14.7 | 52.10 |
| GCA_015559505.1 | D52t1_170925_G1 | 98.32 | 14.0 | 49.60 |
| GCA_015558085.1 | BSD2780120874b_170522_D7 | 98.30 | 14.0 | 49.59 |
| GCA_019129365.1 | MSK.17.32 | 98.28 | 14.0 | 49.58 |
| GCA_023497865.1 | NB2B-16-TSAB | 98.27 | 14.0 | 49.58 |
| GCA_003429385.1 | 6 | 98.21 | 14.0 | 49.55 |
| GCA_003473105.1 | AM13-11 | 98.11 | 14.0 | 49.50 |
| GCA_003467335.1 | AM41-17 | 98.31 | 13.2 | 46.99 |
| GCA_003472095.1 | AM14-37 | 98.30 | 13.2 | 46.98 |
| GCA_015555595.1 | D59t2_181005_G7 | 98.29 | 13.2 | 46.98 |
| GCA_001406735.1 | 2789STDY5608862 | 98.23 | 13.2 | 46.95 |
| GCA_022135325.1 | DFI.7.20 | 98.19 | 13.2 | 46.93 |
| Local | 235B | 98.18 | 13.2 | 46.93 |
| GCA_015558745.1 | 1001713B170214_170313_A6 | 98.18 | 13.2 | 46.93 |
| GCA_003464325.1 | AF15-3 | 98.13 | 13.2 | 46.90 |
| GCA_015554265.1 | 1001099B_141217_F6 | 98.31 | 12.5 | 44.38 |
| GCA_000817995.1 | BBMN23 | 98.29 | 12.5 | 44.37 |
| GCA_018785715.1 | MCC257 | 98.29 | 12.5 | 44.37 |
| GCA_005845205.1 | 1001271st1_A4 | 98.28 | 12.5 | 44.36 |
| GCA_001010915.1 | 150 | 98.24 | 12.5 | 44.35 |
| GCA_002108015.1 | 487B | 98.20 | 12.5 | 44.33 |
| GCA_001406455.1 | 2789STDY5608824 | 98.19 | 12.5 | 44.32 |
| GCA_015549865.1 | D31t1_170403_E3 | 98.09 | 12.5 | 44.28 |
| GCA_014524935.1 | HD17T2H | 98.29 | 11.8 | 41.76 |
| GCA_015553845.1 | BSD2780061687b_171204_D1 | 98.23 | 11.8 | 41.73 |
| Local | 946B | 98.34 | 11.0 | 39.17 |
| GCA_002108165.1 | LMG 18897 | 98.34 | 11.0 | 39.17 |
| GCA_015557685.1 | 1001262B_160229_D10 | 98.34 | 11.0 | 39.17 |
| GCA_002108045.1 | AD2-8 | 98.26 | 11.0 | 39.14 |
| Local | 14B | 98.24 | 11.0 | 39.13 |
| GCA_015558565.1 | 1001275B_160808_D1 | 98.23 | 11.0 | 39.12 |
| GCA_003856735.1 | P2P3 | 98.19 | 11.0 | 39.11 |
| GCA_015548665.1 | 1001270B_150601_E2 | 98.14 | 11.0 | 39.09 |
| GCA_022136605.1 | DFI.5.12 | 97.88 | 11.0 | 38.99 |
| Local | 740B | 98.33 | 10.3 | 36.55 |
| GCA_015547885.1 | BSD2780061688_150302_F11 | 98.30 | 10.3 | 36.54 |
| GCA_001756865.1 | Km 4 | 98.29 | 10.3 | 36.54 |
| GCA_015558575.1 | D46t1_190503_C9 | 98.18 | 10.3 | 36.50 |
| GCA_002108075.1 | AL46-2 | 98.18 | 10.3 | 36.50 |
| Local | 713B | 98.29 | 9.6 | 33.93 |
| GCA_019127685.1 | MSK.21.29 | 98.23 | 9.6 | 33.91 |
| Local | 2419–10B | 98.31 | 8.8 | 31.33 |
| GCA_015553955.1 | 1001095IJ_161003_A7 | 98.23 | 8.8 | 31.30 |
| Local | 2141B | 97.94 | 8.8 | 31.21 |
| GCA_004167585.1 | ca_0067 | 98.31 | 8.1 | 28.71 |
| Local | 75B | 98.29 | 8.1 | 28.71 |
| GCA_015560095.1 | 1001270J_160509_E8 | 98.28 | 8.1 | 28.71 |
| GCA_000737885.1 | 22L | 98.32 | 7.4 | 26.11 |
| GCA_003458805.1 | AF21-27 | 97.94 | 5.9 | 20.80 |
| GCA_003465205.1 | AF14-56 | 98.08 | 5.1 | 18.23 |
| GCA_000702865.1 | DSM 20087 | 97.74 | 2.9 | 10.38 |
Then, an Average × Prevalence index (A × P index) was generated, integrating genetic data produced as average nucleotide identity (ANI) values between de-replicated genetically unique strains and ecological data based on lineage prevalence among metagenomes (19). This procedure allowed the selection of a reference strain from lineage 12, i.e., B. adolescentis PRL2023, with the highest A × P score (98.77), corresponding to the most representative B. adolescentis strain inhabiting the GIT of healthy humans (Table 2). In contrast, the B. adolescentis type strain, i.e., ATCC 15703 showed a much lower A × P value of 54.61. Thus, in silico analyses revealed that from a genomic perspective, PRL2023 can be considered the best representative B. adolescentis strain of the human gut, while this strain is also ecologically significant due to its global distribution among metagenomic samples. Based on these data, the proposed prototype of the B. adolescentis species, strain PRL2023, was further investigated through in silico genomic screenings and in vitro interaction with the intestinal microbiota community.
Genetic features of B. adolescentis PRL2023
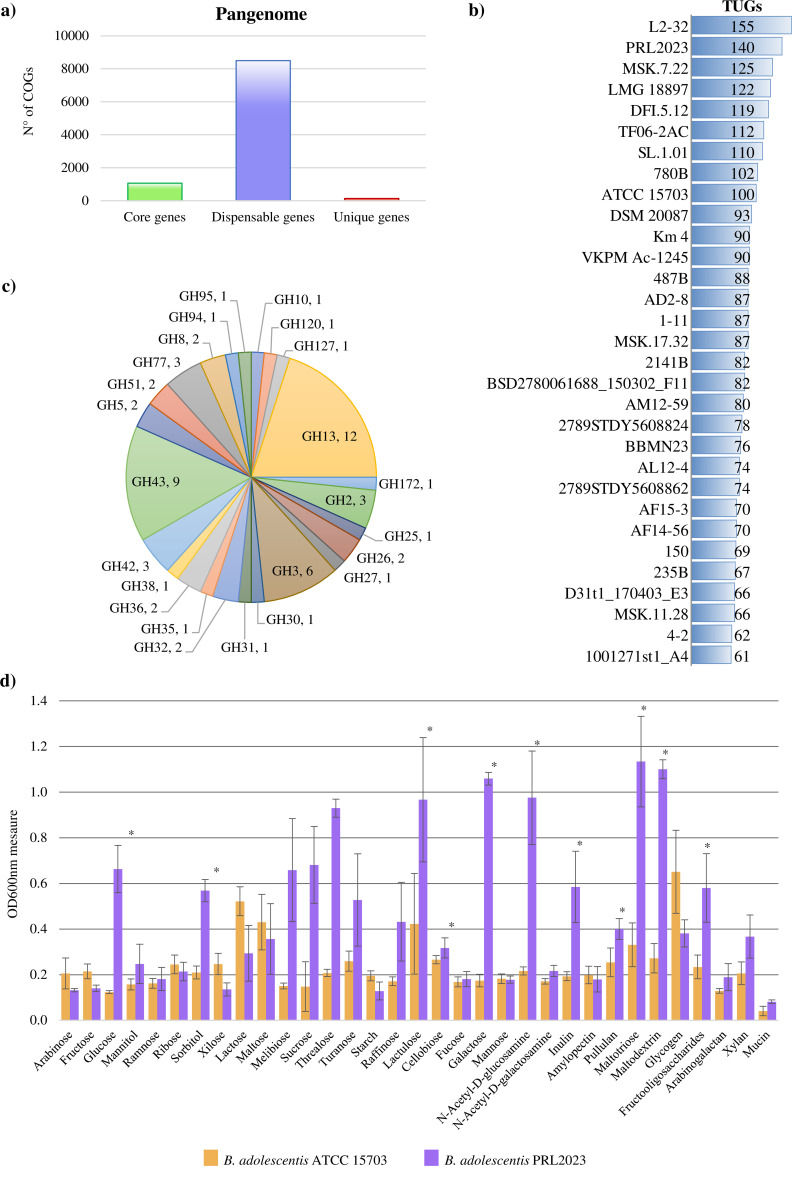
To obtain a comprehensive overview of the genetic traits of PRL2023, its genome sequence was decoded employing a combination of short- and long-read sequencing technologies (see Materials and Methods), resulting in a complete genome sequence (i.e., a single contig representing a circular chromosome). Then, a comparative genomic analysis of the B. adolescentis species was performed, including the predicted proteome of each representative strain of the above-identified 79 lineages (Table 2). Thus, pangenome and core-genome analyses of this taxon were undertaken following a previously described method based on Clusters of Orthologous Groups (COGs) (32–34). This analysis resulted in the identification of 11,024 COGs, representing the pangenome of the species. Among them, 1,068 COGs were shared between all 79 B. adolescentis assessed genomes, thus constituting their core genome (Fig. 1a). Furthermore, truly unique genes (TUGs) of each strain were detected with an average of 63 TUGs per genome. Interestingly, B. adolescentis PRL2023 was revealed to be the second strain with the highest abundance of TUGs when compared to members of the other lineages (Fig. 1b). Among the identified TUGs of strain PRL2023, three putative genes encoding carbohydrate-active enzymes were identified, and we, therefore, decided to further investigate the metabolic capabilities of B. adolescentis PRL2023.
Fig 1.
Genomic analyses and carbohydrate profiling of members of the B. adolescentis taxon. Panel (a) shows the number of core genes (green), unique genes (red), and dispensable genes (blue) identified in the pangenome of the B. adolescentis species. Panel (b) displays the distribution of TUGs among putative B. adolescentis prototypes (only strains with more than 60 TUGs are reported). Panel (c) depicts the glycosyl hydrolase (GH) distribution in PRL2023. Panel (d) exhibits the growth performance of B. adolescentis PRL2023 and ATCC 15703 on different carbohydrates as measured by optical density at 600 nm (OD600nm).
An in silico prediction of the glycobiome of B. adolescentis PRL2023, based on the Carbohydrate-Active enZYmes (CAZy) database (35), revealed that 60 genes were predicted to encode glycosyl hydrolases (GHs) encompassing 24 different families (Fig. 1c). Among them, 12 genes were predicted to belong to the GH13 family, which encompasses enzymes with amylase and pullulanase activities. The high abundance of GH13 in the genome of PRL2023 is in line with a previous investigation of the B. adolescentis glycobiome based on 18 strains that highlighted a rich repertoire of GHs belonging to the GH13 and GH45 families (31). In accordance, members of the latter GH family were represented by the predicted products of nine genes among the PRL2023 proteome. Thus, glycobiome profiling revealed the tendency of PRL2023 to process various polysaccharides such as plant-derived starch or animal-derived glycans like glycogen. Furthermore, members of additional GH families were identified, which are predicted to degrade various glycans, e.g., β-xylosidase (GH3 and GH120), α-L-arabinofuranosidase (GH51), β-L-arabinofuranosidase (GH127), α-galactosidase (GH36), β-galactosidase (GH35 and GH42), α-mannosidase (GH38), and β-mannosidase (GH26). Overall, the predicted glycobiome of PRL2023 indicates a preponderance of this strain to degrade dietary glycans, seemingly supporting the establishment of a symbiotic relationship with humans.
B. adolescentis PRL2023 growth profiles on different carbohydrates
Aiming to validate the activity of the in silico identified carbohydrate-active enzymes, we performed growth experiments of B. adolescentis PRL2023 and the B. adolescentis type strain, i.e., ATCC 15703, in a medium in which different carbohydrates, including both plant- and host-derived glycans, were individually included as the sole carbon source (Fig. 1d). In detail, for growth-profiling experiments, we used a carbohydrate-free basic MRS medium, which was supplemented with one of a collection of 33 different sugars, as the sole carbon source (Table S2; Fig. 1d). Upon Mann-Whitney test with Benjamini–Hochberg correction (cutoff P-value < 0.05), the comparative growth assay showed widespread statistically significant differences in growth performances across the two B. adolescentis strains. In detail, B. adolescentis PRL2023 displays a greater ability to grow on inulin, pullulan, maltotriose, maltodextrin, and fructooligosaccharides (final OD value from 0.4 to 1.1; all Benjamini–Hochberg corrected P-values < 0.05) when compared to the type strain ATCC 15703 (Table S2; Fig. 1d). These data demonstrate that the carbohydrate-metabolizing abilities of B. adolescentis PRL2023 were more extensive when compared to the ATCC 15703 type strain, corroborating our in silico strain-based tracking analyses, which identified PRL2023 as the more prevalent strain among the healthy human gut (Table 2). Moreover, together with plant-derived glycans, PRL2023 shows appreciable growth on different hexose-containing sugars, such as lactulose, cellobiose, galactose, and N-acetyl-D-glucosamine (final OD value from 0.3 to 1.0; all Benjamini–Hochberg corrected P-values < 0.05) (Table S2; Fig. 1d), in accordance with the presence of GHs in its predicted glycobiome for the degradation of simple sugars, i.e., GH26, GH35, GH36, GH38, and GH42. Moreover, both strains are rather limited in their ability to utilize mucin (Table S2; Fig. 1d), in agreement with previous works showing that the ability to use mucin as a carbon source is a feature that has only been described for Bifidobacterium bifidum and B. longum species (31, 36, 37). However, growth profiles indicated that the extensive repertoire of B. adolescentis PRL2023-encoded GHs provides this strain a clear advantage in metabolizing several carbon sources, suggesting that its dominance in most of the human population may at least be partially based on its expansive glycan degradation abilities.
Assessing the gastrointestinal resilience of PRL2023 in vitro
To colonize their natural ecological niche, bifidobacteria are expected to counteract the adverse and hostile environmental conditions that they encounter during transit toward the large intestine and in the colon, including exposure to biliary salts, osmotic stress, or the extreme acidic conditions encountered during their passage through the stomach (1, 38). To evaluate the ability to survive osmotic stress or exposure to bile salts, B. adolescentis PRL2023 was exposed to different concentrations of NaCl (2%, 6%, and 10%) and Oxgall (0.5%, 1%, and 2%) for 3 h (39), after which cell viability was monitored through flow cytometry. Interestingly, PRL2023 showed a survival rate of 97.23% at 2% NaCl, which is in line with those observed for other gut commensal bifidobacterial strains (40), indicating that this strain is rather tolerant to osmotic stress (Table 3). Exposure to bile salts is another harsh condition that bifidobacteria have to cope with in the GIT. It is widely reported in the literature that the physiological bile salt concentration in the human gut roughly varies from 0.3% to 0.4% (41). Interestingly, PRL2023 showed a survival rate of 30.9% at a concentration higher than the physiological one of Oxgall, i.e., 0.5%. In addition, we also tested the ability of PRL2023 to survive in an acidic environment, simulating the human stomach. In general, bifidobacterial cells have been reported to display low viability toward acidic environment with values between 41.8% and 79.3% at the typical pH 2 of the stomach (39, 40, 42–44). Specifically, PRL2023 cells showed excellent survival of 78.3% following 2 h incubation at pH 4. However, much lower survival rates were observed when the acid challenge assays were done at pH 2.0 and pH 3.0 (Table 3). Accordingly, in vitro survival assays of the selected Bifidobacterium prototype, mimicking harsh GIT conditions, highlighted how this strain can tolerate stressful conditions typical of the intestinal environment.
TABLE 3.
Viability of B. adolescentis PRL2023 when exposed to human gastrointestinal challenges
| Viablea PRL2023 cells (%) | Non-viablea PRL2023 cells (%) | |
|---|---|---|
| MRS 3h | 96.89 | 3.11 |
| 2% NaCl | 97.23 | 2.77 |
| 6% NaCl | 39.35 | 60.65 |
| 10% NaCl | 50.67 | 49.33 |
| 0.5% Bile salts | 30.94 | 69.06 |
| 1% Bile salts | 9.42 | 90.58 |
| 2% Bile salts | 11.14 | 88.86 |
| MRS 2h | 75.13 | 24.87 |
| pH 2.0 | 12.21 | 87.79 |
| pH 3.0 | 11.27 | 88.73 |
| pH 4.0 | 78.26 | 21.74 |
Data are expressed as the average of the obtained triplicates.
In addition, to investigate the ability of this strain to interact with the host, we evaluated the adhesive performance of B. adolescentis PRL2023 and those of the ATCC 15703 strain, which is the type strain of B. adolescentis, to the human intestinal mucosal cells by using a previously described methodology (45–47). Interestingly, both strains were able to adhere to HT29-MTX cell monolayers, as demonstrated by the adhesion index of 89,333 ± 4 and 86,400 ± 9 determined for PRL2023 and ATCC 15703 cells, respectively (Fig. S2). Several factors can influence the adhesion of a bacterial strain, including culture conditions and HT29-MTX cell characteristics, so there is not an “ideal value” of the adhesion index to be used as a reference. For this reason, the experimental approach was also applied to the Bifidobacterium animalis subsp. lactis Bb-12 strain (45), which is commonly used as a probiotic supplement. The comparison revealed an adhesion index of Bb-12 less than 20,000 (Fig. S2), highlighting a clear-cut superior adhesion of both B. adolescentis PRL2023 and the type strain ATCC 15703. In addition, an adhesion assay on mucin was performed (48), highlighting a higher relative adhesion to mucin of PRL2023 (61.5%) when compared to B. adolescentis ATCC 15703 (51.7%). These results are consistent with our other observations and indicate that the prototype PRL2023 strain is ecologically adapted to the human gut microbiota and likely expresses genes that are predicted to play a role in enhancing the colonization of the human intestinal mucosa. The improved capacity of the strain to withstand challenging gut environment conditions and its capacity to bind to the human epithelial cells imply that PRL2023 is endowed with genetic features able to support its potential exploitation as a novel probiotic microorganism.
Investigation of the molecular interaction between PRL2023 and common members of the human gut microbiota
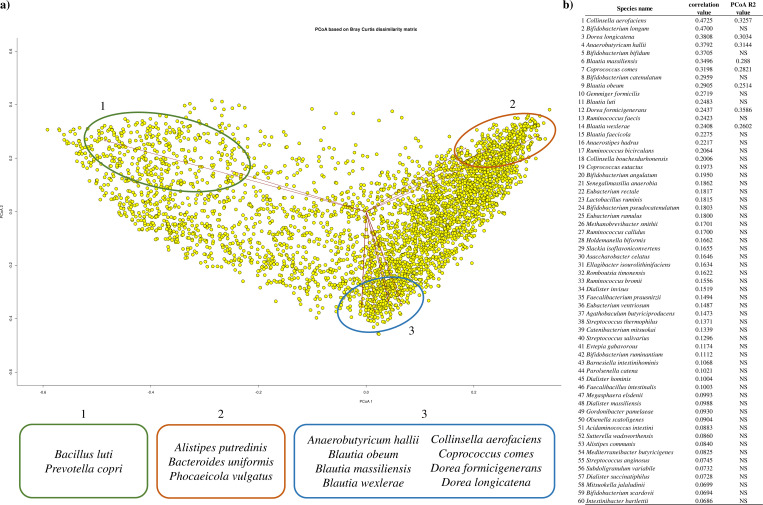
A correlation analysis among bacterial species inhabiting the human gut microbiota of adults allowed us to identify those species that commonly share the same ecosystem with B. adolescentis taxon. Overall, 88 microbial species highlighted a positive and significant correlation with B. adolescentis (Benjamini–Hochberg, false discovery rate, FDR P-values < 0.05) (Table S3). Among bifidobacteria, B. adolescentis species was found second only after B. longum species, which revealed a significant positive correlation with 91 human gut bacteria (Table S3). A principal coordinate analysis (PCoA) was performed to further investigate which bacterial species were most important in terms of their association with human gut microbiota variability (Fig. 2a). Normalized data highlighted that among the 88 bacterial species that positively correlated with the B. adolescentis taxon, only eight were found significant in defining the gut microbiome variability (R2 > 0.2, FDR P-values < 0.005), i.e., Collinsella aerofaciens, Dorea longicatena, Anaerobutyricum hallii, Blautia massiliensis, Coprococcus comes, Blautia obeum, Dorea formicigenerans, and Blautia wexlerae. Interestingly, the latter species is one of the 14 species with the highest correlation values with respect to the B. adolescentis taxon (Fig. 2b), highlighting a clear interconnection between bacteria shaping the diversity of the human gut microbiota.
Fig 2.
B. adolescentis and its role in shaping the human microbiome. Panel (a) shows a PCoA where each yellow circle represents a human microbiome included in the analysis. Significant microorganisms in shaping the variability of the human gut are reported below the PCoA (R2 > 0.2, FDR P-values < 0.005). Panel (b) displays the list of significant bacteria correlating with B. adolescentis. NS, not significant.
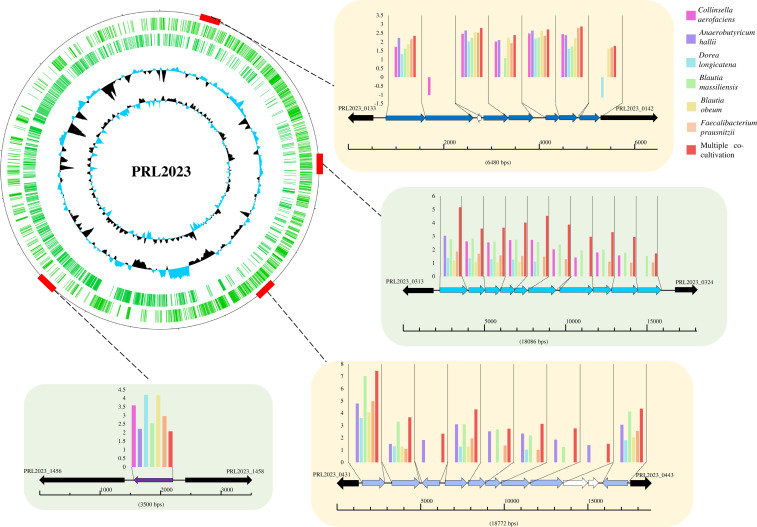
In order to evaluate the interactions between the proposed prototype B. adolescentis PRL2023 strain and a selected number of bacterial species that shape the human gut microbiota of adults, we performed co-cultivation assays with PRL2023. In detail, we performed six co-cultivation assays where B. adolescentis PRL2023 cells were co-incubated with cells harboring Collinsella aerofaciens DSM 3979, Dorea longicatena DSM 13814, Anaerobutyricum hallii DSM 3353, Blautia massiliensis DSM 101187, Blautia obeum DSM 25238, and Faecalibacterium prausnitzii DSM 107838. The latter species was chosen in addition to the most relevant bacteria that co-occurred with B. adolescentis due to its notable scientific interest with respect to gut microbiota and associated host health (49–51). Furthermore, an additional co-cultivation assay involving all of the above-mentioned species together with PRL2023 was performed. A quantitative PCR (qPCR) approach was used to quantify the bacterial DNA of each species to the total DNA extracted from co-cultivation experiments. This qPCR analysis highlighted that the PRL2023 strain can grow in all the co-culture experiments with the different gut microbial commensals (Fig. S1). Thus, shotgun metatranscriptomic was carried out in each co-cultivation assay to investigate how PRL2023 interacts with other commensals. This analysis revealed that, compared to the reference condition (PRL2023 grown as a monoculture), the number of PRL2023 upregulated genes was always higher than downregulated genes, with a ratio ranging from 0.84 to 0.15 (down-/upregulated genes), in Anaerobutyricum hallii and Blautia obeum co-cultivation, respectively (Table S4). Particularly, PRL2023 cells, when cultivated with other intestinal commensals, were shown to elicit increased transcription of genes belonging to a tight adherence pilus locus (PRL2023_0134-0141), i.e., seven genes encoding Type IV pilus assembly and secretion proteins (Fig. 3). In this context, it is well known that pili are considered one of the principal structures involved in microbe-host interactions (1, 45, 52) (Fig. 3). Thus, metatranscriptomics data demonstrate that other human commensals directly enhanced the host interaction ability of PRL2023. Furthermore, the expression of additional genes illustrated the synthesis of extracellular structures involved in interactions, such as a pilus assembly protein, a cell wall anchor domain-containing protein, and a sortase putatively involved in pilin biosynthesis (Table S4), corroborating the notion that PRL2023 is actively transcribing such complexes in co-culture.
Fig 3.
Transcriptome analyses of PRL2023 when co-cultivated with other human gut commensals. The circular genome atlas represents the collection of genes belonging to B. adolescentis PRL2023 (blue circles). Internal circles illustrate PRL2023 GC% deviation and GC skew (G − C/G + C), while the external maps highlight loci with increased transcription (compared to mono-cultivation control) as identified in co-cultivation assays. Each arrow indicates an open reading frame (ORF), whereas the length of the arrow is proportional to the length of the predicted ORF. Histograms upon each ORF report the expression with respect to the control condition, and the Y axis reports normalized count reads (TMM).
Additionally, genes predicted to be involved in carbohydrate metabolism and their uptake were upregulated in co-cultivation assays with other microorganisms. Recently, it has been shown that the presence of other microbes allowed the development of cross-feeding strategies, in which bifidobacteria access a wider set of carbon sources compared to cells grown in monoculture (20, 53) (Fig. 3). Here, metatranscriptomics data revealed that two loci, in particular, increased their transcription under such co-cultivation circumstances, unveiling the activation of genes involved in the degradation of a complex polysaccharide, i.e., xylan (PRL2023_0314-0323), and a simple monosaccharide, i.e., xylose (PRL2023_0432-0442), that are typical indigestible pentose sugars found in the human diet (Fig. 3). The structure of both loci unveiled the presence of two dedicated uptake systems as well as degradative carbohydrate enzymes (Table S4), highlighting the activation of a metabolic machinery for the uptake and degradation of xylose and xylose-based oligosaccharides. As previously reported, xylose-containing glycans are metabolized in multiple associations due to the rapid depletion of simple substrates (54). Thus, resource competition in the bi-association assays, due to the combination of GH enzymes of other gut commensals, may have activated a plant-derived carbohydrate metabolism that was not expressed in mono-association cultures, a phenomenon that has also been observed previously (55, 56). Notably, the ability to switch carbohydrate metabolism between different substrates may represent successful strategies allowing this PRL2023 strain to colonize and/or persist in the gut microbiota of adults. Furthermore, additional enzymes involved in the break down of complex plant carbohydrates were identified to be upregulated in co-cultivation assays as well as other predicted β-xylanase-encoding genes scattered across the genome (Table S4). In this context, PRL2023 showed the enhanced transcription of genes predicted to encode α- and β-glucosidases, α- and β-galactosidases, β-mannosidases, and β-fructofuranosidases (Table S4), together with synthase-encoding genes for the conversion of glucose into glycogen for storage.
Interestingly, a gene encoding a hypothetical protein of PRL2023, i.e., PRL2023_1457, was overexpressed in any of the bi-association experiments and when PRL2023 was co-cultivated with all these intestinal commensals (Fig. 3). Specifically, sequence homology analyses revealed the presence of a DUF4192 motif, suggesting its involvement in conjugation and DNA metabolism (57). The expression of this unknown gene may be pivotal in understanding the interaction between PRL2023 and (members of) the human microbiota.
Conclusions
To date, a large body of scientific literature indicates that bifidobacteria are well known for the benefits exerted on their host, such as promoting the intestinal barrier integrity, improving gut homeostasis, and supporting the development of the host immune system (2, 16, 18, 58, 59). In this context, B. adolescentis stands out among the most abundant species of bifidobacteria residing in the human adult intestine (11, 60). In the current study, an ecological and phylogenomic-driven system was applied allowing the identification of B. adolescentis PRL2023 strain as the most representative taxon of this species inhabiting the human gastrointestinal tract of adults. As recently reported, the use of a suitable strain for experimental research is necessary to manipulate bacteria sharing the most representative genomic features found in nature (19, 27).
Comparative genome analysis, involving the prototype PRL2023 and other B. adolescentis strains, revealed a high number of unique genes, including genes involved in the degradation of complex sugars typical of an adult host diet. Moreover, in vitro experiments involving models that simulate typical harsh gastrointestinal tract conditions as well as contact with human cell lines suggest that PRL2023 can survive well in the intestinal environment and efficiently adhere to the human epithelium. Thus, based on our data, the selected strain should be able to reach and colonize the intricate human intestinal ecological niche. In addition, metatranscriptomics analyses based on PRL2023 in correlation with other intestinal microbes, enhanced our understanding as to why this prototype is widely distributed among the population, highlighting the ability of PRL2023 to interact with its host as well as its ability to switch its carbohydrate metabolism to effectively compete with other members of the human microbiota. Ultimately, to further advance our scientific knowledge regarding PRL2023 and to validate our in vitro observations on its abilities to interact with the human host, a human clinical trial involving this strain will be necessary. Nonetheless, our reported data suggest that this strain represents a very promising candidate whose potential health-promoting activities are worthy to be determined.
MATERIALS AND METHODS
Metagenome data set selection
In this project, 4,019 publicly available human gut microbiome data sets were retrieved from 82 cohorts of healthy adults (age from 18 to 80 years) (Table S1). Additionally, 76 samples belonging to healthy elderly (age > 80 years) were used to explore the aging-associated microbial diversity (Table S1).
Taxonomic classification of metagenomic reads
To analyze high-quality DNA sequence data, each data set was subjected to a filtering step removing low-quality reads (minimum mean quality score 20, window size 5 nt, quality threshold 25, and minimum length 100 nt) using the fastq-mcf script (https://github.com/ExpressionAnalysis/ea-utils/blob/wiki/FastqMcf.md). Filtered reads were then collected and taxonomically classified through the METAnnotatorX2 pipeline (61), using the up-to-date RefSeq (genome) database retrieved from the NCBI (https://www.ncbi.nlm.nih.gov/refseq/). Short-read sequences were taxonomically classified based on their sequence identity using megablast (62).
B. adolescentis prototype selection
Complete and partial genomes of 113 B. adolescentis strains were retrieved from the RefSeq NCBI database representing the collection of validated publicly sequenced genomes of this taxon. By selecting strains for in silico analyses, we admittedly ignored metagenomic assembled genomes, so as to investigate previously cultivated bacteria only. The genome sequence of the reference strain ATCC 15703 was used to discard strains showing an average nucleotide identity lower than 94% employing the software fastANI (63) to exclude taxonomically misclassified microorganisms. Furthermore, genome sequence quality was estimated for completeness and contamination using CheckM (64). High-quality genomes were then subjected to a de-replication-based analysis aiming to reduce strain redundancy among bifidobacterial genome sequences using dRep v2.0 (65). Among strains displaying sequence identity >99.8%, a single reference genome was selected for further analysis, representing putative prototypes of the B. adolescentis species. Then, a k-mer-based analysis to explore the distribution of each putative prototype was investigated using InStrain software with a k-mer size of 23 (19). The selection of the B. adolescentis prototype of the healthy human gut was based on a previously validated index called A × P (19), defined as (the average ANI value of genomes constituting the same clade) × (prevalence score of the strain in the data set) × (100).
Genome sequencing
The genome sequence of PRL2023 was determined by GenProbio Srl (Parma, Italy) using the MiSeq platform (Illumina, UK). Genome libraries were prepared using an Illumina Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, CA, USA). Libraries were quantified using a fluorometric Qubit quantification system (Life Technologies, USA), loaded on a 2200 TapeStation instrument (Agilent Technologies, USA), and normalized to 4 nM. Sequencing was performed using the Illumina MiSeq platform with a 600-cycle flow cell version 3 (Illumina Inc., San Diego, CA, USA). Additionally, PRL2023-extracted DNA was subjected to whole-genome sequencing using the Nanopore DNA sequencing platform according to the supplier’s protocol (Oxford Nanopore, UK).
Genome assembly
Long reads were filtered by quality using the Filtlong tool (https://github.com/rrwick/Filtlong), while short reads were filtered through the fastq-mcf script (https://github.com/ExpressionAnalysis/ea-utils). Filtered fastq files of Nanopore long reads obtained from genome sequencing efforts were then used as input for genome assembly through CANU software (66). The resulting genome sequence has been polished through Polypolish (67) using Illumina paired-end reads. The whole process was managed by the MEGAnnotator2 pipeline (68).
Comparative genomics
Open reading frames of each B. adolescentis genome were predicted with Prodigal (69) and annotated utilizing the MEGAnnotator2 pipeline (68, 70).
Proteomes were employed for a pangenome calculation using the PGAP (71), to identify orthologs between analyzed B. adolescentis strains byBLAST analysis (cutoff E value < 1 × 10−5; 50% identity over at least 80% of both protein sequences). The resulting output was then clustered into protein families named COGs through graph theory-based Markov clustering algorithm, using the gene family method. Pangenome profiles were built using an optimized algorithm incorporated in the PGAP software, based on a presence/absence matrix that included all identified COGs in the 79 analyzed genomes.
Glycobiome profiling
The proteome of PRL2023 was screened for genes predicted to encode carbohydrate-active enzymes based on sequence similarity to genes classified in the CAZy database (35). Thus, each gene sequence was screened for orthologs against the dbCAN2 meta server (72) composed of 2,141,452 coding sequences using HMMER v3.3.2 (cutoff E-value of 1 × 10−15 and coverage > 0.35) and DIAMOND (E-value <1 × 10−102).
Bacterial cultivation conditions
B. adolescentis PRL2023 was cultivated in the de Man-Rogosa-Sharpe (MRS) medium (Sharlau Chemie, Spain) supplemented with 0.05% (wt/vol) L-cysteine hydrochloride (Merk, Germany) and incubated at 37°C in a chamber (Concept 400, Ruskinn) with an anaerobic atmosphere (2.99% H2, 17.01% CO2, and 80% N2). For bi-association experiments, Collinsella aerofaciens DSM 3979, Dorea longicatena DSM 13814, Anaerobutyricum hallii DSM 3353, Blautia massiliensis DSM 101187, Blautia obeum DSM 25238, and Faecalibacterium prausnitzii DSM 107838 were obtained from DSMZ-German Collection of Microorganisms and Cell Cultures GmbH. These microorganisms were cultivated anaerobically in yeast extract-casein hydrolysate-fatty acid (YCFA) medium in Hungate tubes at 37°C for 48 h.
pH, sodium chloride, and bile salt tolerance tests
To evaluate the ability of the selected strains to tolerate various pH levels, B. adolescentis PRL2023 was cultivated in 10 mL of MRS broth at 37°C under anaerobic conditions to reach a final concentration of 108 cells/mL. Subsequently, cells were centrifuged at 3,000 rpm for 8 min, washed with phosphate-buffered saline (PBS, pH 6.5), and resuspended in 10 mL of MRS broth whose pH was adjusted to 2.0, 3.0, or 4.0 with the addition of HCl. Cells were incubated under anaerobic conditions at 37°C for 2 h, as previously described (39). The same procedure was performed to assess the ability of bifidobacteria to tolerate different concentrations/levels of NaCl (2%, 6%, and 10%) or bile salts (Sigma Aldrich, USA) (0.5%, 1%, and 2%) with an exposure of 3 h to these stressful conditions, as previously reported (39). All experiments were carried out in triplicate, and a control sample was obtained by inoculating bifidobacterial cells in MRS broth. After incubation, cell viability was evaluated using the LIVE/DEAD BacLight Bacterial Viability kit (ThermoFisher Scientific, USA) and an Attune NxT flow cytometer (ThermoFisher Scientific, USA).
Flow cytometry bacterial viability assay
Following exposure to acidic environment, or various bile salts or NaCl concentrations, a 10-fold serial dilution in PBS was obtained from each tested condition. The diluted cells were then used for a flow cytometry cell viability assay using the fluorescent dyes SYTO9 (3.34 mM) and PI (20 mM) of the LIVE/DEAD BacLight Bacterial Viability kit (ThermoFisher Scientific, USA), following the manufacturer’s protocol (Manual of the LIVE/DEAD BacLight Bacterial Viability and counting kit, ThermoFisher Scientific, USA). Briefly, two aliquots of 1 mL of bacterial cell dilution (1:1,000) were harvested by centrifugation at 3,000 rpm for 8 min and washed with PBS. Subsequently, one of the two aliquots of bacterial suspension was exposed to 70% isopropyl alcohol and kept on ice for 1 h to permeabilize cell membranes and induce cell death, while the other 1 mL aliquot was maintained in PBS to preserve cell viability. Subsequently, 1.5 µL of a specific dye was added to samples for the single staining assay, while for the double staining assay, 1.5 µL of both dyes was added to the samples. Once stained, samples were incubated in the dark for 15 min at room temperature. Furthermore, while single-stained controls were used for instrument parameter adjustment, non-stained cells were used as a background control. Cell viability assays were performed with the Attune NxT flow cytometer (ThermoFisher Scientific, USA), and obtained data sets were analyzed with the Attune NxT flow cytometer software.
Carbohydrate-dependent growth assays
To validate in silico findings, we performed growth assays on multiple carbon sources involving the prototype PRL2023 as well as the type strain B. adolescentis ATCC 15703 as reference. Notably, in silico analyses performed in this study generated predictions with regard to the (carbohydrate) metabolic abilities of the above-mentioned strains and were further described in the Results and Discussion section. B. adolescentis strains were cultivated overnight on a semisynthetic MRS medium supplemented with 0.05% (wt/vol) L-cysteine hydrochloride at 37°C under anaerobic conditions. Subsequently, cells were diluted in MRS without glucose to obtain an OD600 nm ~ 1, and 15 µL of the diluted cells was inoculated in 135 µL of MRS without glucose supplemented with 1% (wt/vol) of a particular sugar in a 96-well microtiter plate and incubated in an anaerobic cabinet. Specifically, each carbohydrate was dissolved in MRS without glucose previously sterilized by autoclaving at 121°C for 15 min. Subsequently, each obtained solution was filter sterilized using a 0.2 µm filter size prior to use. Cell growth was evaluated by monitoring the optical density at 600 nm using a plate reader (Biotek, VT, USA). The plate was read in discontinuous mode, with absorbance readings performed at 3-min intervals for three times after 48 h of growth, and each reading was ahead of 30 s of shaking at medium speed. Cultures were grown in triplicates, and the resulting growth data were expressed as the average of these replicates. Carbohydrates tested in this study were purchased from Merck (Germany) and were reported in Table S2.
In vitro evaluation of the interaction of PRL2023 cells with selected members of the human gut microbiota
To evaluate how B. adolescentis PRL2023 interacts with other gut microbial players, batch cultures were set up to co-cultivate the selected strain with six different intestinal commensals, i.e., Collinsella aerofaciens DSM 3979, Dorea longicatena DSM 13814, Anaerobutyricum hallii DSM 3353, Blautia massiliensis DSM 101187, Blautia obeum DSM 25238, and Faecalibacterium prausnitzii DSM 107838. For bi-association experiments, overnight cultures of each microorganism were diluted to obtain an approximate OD value of 1.0, as previously described (73). Each culture was inoculated at 0.1% (vol/vol) into YCFA medium (74–76). We performed six different experiments in which B. adolescentis PRL2023 was inoculated, respectively, with six different intestinal players mentioned above and one experiment where all microorganisms were cultivated altogether. Batch cultures were performed in triplicate and incubated under anaerobic conditions and in Hungate tubes at 37°C. After 8 h of incubation, cultures were centrifuged at 7,000 rpm for 5 min, the supernatants were discarded, while the obtained bacterial pellets were used for RNA extraction (53, 77, 78). Moreover, pellets were subjected to DNA extraction using the GeneElute bacterial genomic DNA kit (Sigma, Germany) following the manufacturer’s instructions. Each sample was subjected to a different cycle of qPCR using strain-specific primers: B0703_0097_FW(5′-TGCAATGATGAATCCACGCC-3′) and B0703_0097_RV (5′-GCGGTTGAACTCGAACAGAT-3′) for B. adolescentis PRL2023. qPCR was performed using qPCR green master mix (PowerUp SYBR Green Master Mix for qPCR, ThermoFisher Scientific, USA) on a CFX96 system (Bio-Rad, CA, USA) following previously described protocols (79, 80). PCR products were detected with SYBR green fluorescent dye and amplified according to the following protocol: one cycle of 50°C for 2 min, followed by one cycle of 95°C for 2 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. The melting curve was 65°C–95°C with increments of 0.5°C/s. In each run, negative controls (no DNA) were included. A standard curve was built using the CFX96 software (Bio-Rad).
RNA extraction
Total RNA from bacterial cells was isolated using a previously described method (54, 81). Briefly, bifidobacterial cell pellets were resuspended in 1 mL of QIAzol lysis reagent (Qiagen, Germany) in a sterile tube containing glass beads. Cells were lysed by alternating 2 min of stirring the mix on a bead beater with 2 min of static cooling on ice. These steps were repeated three times. Lysed cells were centrifuged at 12,000 rpm for 15 min, and the upper phase was recovered. Bacterial RNA was subsequently purified using the RNeasy Mini Kit (Qiagen, Germany) following the manufacturer’s instructions. Then, the RNA concentration and purity were evaluated using a spectrophotometer (Eppendorf, Germany).
mRNA sequencing analysis
Total bacterial RNA (from 100 ng to 1 µg) was treated to remove rRNA using the QIAseq FastSelect–5S/16S/23S following the manufacturer’s instructions (Qiagen, Germany). The yield of rRNA depletion was checked using a 2200 TapeStation (Agilent Technologies, USA). Then, a whole transcriptome library for prokaryotic RNA was constructed using the TruSeq Stranded mRNA Sample Preparation Kit (Illumina, San Diego, USA). Samples were then loaded onto a NextSeq high-output v2 kit (150 cycles) (Illumina) as indicated by the technical support guide. The obtained reads were filtered to remove low-quality reads using fastq-mcf tool (minimum mean quality 20 and minimum length 100 bp) as well as any remaining ribosomal locus-encompassing reads (61). The retained reads were then aligned to the complete, closed PRL2023 genome sequence through Bowtie2 software (82). Subsequently, quantification of reads mapped to individual transcripts was achieved through the htseq-counts script of HTSeq software in “union” mode (83). Raw counts were then normalized using the trimmed mean of M-values (TMM) method implemented in the EdgeR package (version 3.6.1) [(84) and Log2 fold change (logFC) was used to evaluate the differences in gene expression of PRL2023 cultivated alone (reference condition) and in bi- or multi-associations (test conditions)]. EdgeR package was also used to identify differentially expressed genes at a FDR of 5% and minimal logFC 1.
Mucin adhesion assay of B. adolescentis PRL2023
The effect of bifidobacterial adhesion on mucin was assessed by adapting the protocol described by Valeriano et al. (48). Briefly, 100 µL of a 1 mg mL−1 sterile mucin dissolved in PBS (pH 7.4) was aliquoted into a 96-well microtiter plate (Sarstedt, Germany) and incubated overnight at 4°C. Subsequently, each well was washed with 200 µL of PBS, rinsed, filled with 100 µL of a 20 mg mL−1 sterile bovine serum albumin solution, and incubated at 4°C for 2 h. Two B. adolescentis strains, i.e., PRL2023 and ATCC 15703, were grown at 37°C under anaerobic conditions (2.99% H2, 17.01% CO2, and 80% N2) (Concept 400; Ruskin) in MRS broth (Sharlau Chemie, Barcelona, Spain) supplemented with 0.05% (wt/vol) L-cysteine HCl. Bifidobacterial growth was monitored until a concentration of 108 CFU mL−1 was reached. A total of 100 µL of a corresponding bacterial suspension, previously washed and resuspended in PBS, was added in each well and incubated under anaerobic conditions at 37°C for 1 h. After incubation, each well was washed three times with 200 µL of PBS to remove unbound bacteria. Then, 200 µL of 0.5% (vol/vol) Triton X-100 was added and incubated at room temperature for 2 h, with gentle agitation to detach the adherent bacteria. The viable cell count expressed as CFU mL−1 was determined in all cases by plating on MRS medium. Each assay was performed in triplicate. Percentage adhesion was calculated as follows:
Adhesion of B. adolescentis PRL2023 to HT29-MTX cells
Bifidobacterial adhesion to HT29-MTX cells was assessed following the protocol described by Serafini et al. (40, 47) Briefly, human colorectal adenocarcinoma HT29-MTX cells (kindly provided by Professor A. Baldi, University of Milan) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 µg/mL streptomycin, and 100 U/mL penicillin and maintained in standard culture conditions. For the experiments, HT29-MTX cells were seeded on microscopy cover glasses previously settled into 10 cm2 petri dishes. Confluent cells were carefully washed twice with PBS before bacterial cells were added. B. adolescentis strains, i.e., PRL2023 and ATCC 15703, and B. animalis subsp. lactis BB12 were grown as previously described until a concentration of 5 × 107 CFU mL−1 was reached. The strains were then centrifuged at 3,000 rpm for 8 min, resuspended in PBS (pH 7.3), and incubated with monolayers of HT29-MTX cells. After 1-h incubation at 37°C, cultures were washed twice with 2 mL of PBS to remove unbound bacteria. The cells were then fixed with 1 mL of methanol and incubated for 8 min at room temperature. Cells were then stained with 1.5 mL of Giemsa stain solution (1:20) (Sigma-Aldrich, Milan, Italy) and left in the dark for 30 min at room temperature. After two washes with 2 mL of PBS, the cover glasses were removed from the Petri plate, mounted on a glass slide, and examined using a phase-contrast microscope Zeiss Axiovert 200 (objective, 100×/1.4 oil). Adherent bacteria in 20 randomly selected microscopic fields were counted and averaged. The proportion of bacterial cells that remained attached to the HT29-MTX monolayer was determined to reflect the extent of specific host-microbe interaction. The adhesion index represents the average number of bacterial cells attached to 100 HT29-MTX cells (45–47). A non-parametric Mann-Whitney test was applied for the detection of statistically significant differences. All assays were performed at least in triplicate.
Statistical analysis
Similarities between samples (beta diversity) were calculated by the Bray-Curtis dissimilarity matrix based on species abundance, using the “vegdist” function on RStudio (http://www.rstudio.com/). Beta diversity was represented through PCoA using the function “ape” of the R suite package (85). Moreover, the various detected bacterial species were tested and plotted on the PCoA using the “envfit” and “plot” functions from vegan through R-studios (http://www.rstudio.com/). PERMANOVA analyses were performed on RStudio using 999 permutations to estimate P-values for population differences in PCoA analyses with the adonis2 package. Furthermore, a correlation analysis between the available metadata and the various detected bacterial species of all samples was performed through Spearman’s rank correlation coefficient using the “rcorr” function (https://CRAN.R-project.org/package=Hmisc), and only results that were significantly different from a statistical perspective were retained. The FDR correction is applied to all statistical analyses based on Benjamini and Hochberg correction through the “p.adjust” function (86).
ACKNOWLEDGMENTS
We thank GenProbio Srl for the financial support from the Laboratory of Probiogenomics. Part of this research was conducted at the High-Performance Computing facility of the University of Parma.
D.v.S. is a member of The APC Microbiome Institute funded by the Science Foundation Ireland (SFI) through the Irish Government’s National Development Plan (Grant numbers SFI/12/RC/2273a and SFI/12/RC/2273b). F.T. was funded by the Italian Ministry of Health through Bando Ricerca Finalizzata (Grant Number GR-2018-12365988). C.A. is supported by Fondazione Cariparma, Parma, Italy.
Contributor Information
Marco Ventura, Email: marco.ventura@unipr.it.
Knut Rudi, Norwegian University of Life Sciences, Ås, Norway.
DATA AVAILABILITY
Raw sequences of metatranscriptomics experiments were accessible through the SRA study BioProject PRJNA1011847. Furthermore, the updated genome sequence of B. adolescentis PRL2023 was deposited in the GenBank database with the NCBI RefSeq accession code CP133648.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/aem.02014-23.
Quantitative PCR evaluation of the numerical load of B. adolescentis PRL2023.
Adhesion of B. adolescentis PRL2023 and ATCC 15703 and B. animalis subsp. lactis Bb-12 cells to HT29-X cells monolayers.
Supplemental tables.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Alessandri G, van Sinderen D, Ventura M. 2021. The genus Bifidobacterium: from genomics to functionality of an important component of the mammalian gut microbiota running title: bifidobacterial adaptation to and interaction with the host. Comput Struct Biotechnol J 19:1472–1487. doi: 10.1016/j.csbj.2021.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alessandri G, Ossiprandi MC, MacSharry J, van Sinderen D, Ventura M. 2019. Bifidobacterial dialogue with its human host and consequent modulation of the immune system. Front Immunol 10:2348. doi: 10.3389/fimmu.2019.02348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. 2015. Role of the normal gut microbiota. World J Gastroenterol 21:8787–8803. doi: 10.3748/wjg.v21.i29.8787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tarracchini C, Viglioli M, Lugli GA, Mancabelli L, Fontana F, Alessandri G, Turroni F, Ventura M, Milani C. 2022. The integrated probiotic database: a genomic compendium of bifidobacterial health-promoting strains. MRR 1:9. doi: 10.20517/mrr.2021.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arboleya S, Watkins C, Stanton C, Ross RP. 2016. Gut bifidobacteria populations in human health and aging. Front Microbiol 7:1204. doi: 10.3389/fmicb.2016.01204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Milani C, Ticinesi A, Gerritsen J, Nouvenne A, Lugli GA, Mancabelli L, Turroni F, Duranti S, Mangifesta M, Viappiani A, Ferrario C, Maggio M, Lauretani F, De Vos W, van Sinderen D, Meschi T, Ventura M. 2016. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep 6:25945. doi: 10.1038/srep25945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C, van Sinderen D, Ventura M. 2016. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol 82:980–991. doi: 10.1128/AEM.03500-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol 20:467–476. doi: 10.1016/j.tim.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 9. Turroni F, van Sinderen D, Ventura M. 2021. Bifidobacteria: insights into the biology of a key microbial group of early life gut microbiota. MRR 1:2. doi: 10.20517/mrr.2021.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martin AJM, Serebrinsky-Duek K, Riquelme E, Saa PA, Garrido D. 2023. Microbial interactions and the homeostasis of the gut microbiome: the role of Bifidobacterium. Microbiome Res Rep 2:17. doi: 10.20517/mrr.2023.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes S, Mancabelli L, Lugli GA, Rodriguez JM, Bode L, de Vos W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2017. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev 81:e00036-17. doi: 10.1128/MMBR.00036-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Turroni F, Rizzo SM, Ventura M, Bernasconi S. 2022. Cross-talk between the infant/maternal gut microbiota and the endocrine system: a promising topic of research. Microbiome Res Rep 1:14. doi: 10.20517/mrr.2021.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ventura M, van Sinderen D, Turroni F. 2022. New research frontiers pertaining to the infant gut microbiota. Microbiome Res Rep 1:24. doi: 10.20517/mrr.2022.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ladeira R, Tap J, Derrien M. 2023. Exploring Bifidobacterium species community and functional variations with human gut microbiome structure and health beyond infancy. Microbiome Res Rep 2:9. doi: 10.20517/mrr.2023.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marasco G, Cremon C, Barbaro MR, Stanghellini V, Barbara G. 2022. Gut microbiota signatures and modulation in irritable bowel syndrome. MRR 1:11. doi: 10.20517/mrr.2021.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hidalgo-Cantabrana C, Delgado S, Ruiz L, Ruas-Madiedo P, Sánchez B, Margolles A. 2017. Bifidobacteria and their health-promoting effects. Microbiol Spectr 5. doi: 10.1128/microbiolspec.BAD-0010-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bottacini F, van Sinderen D, Ventura M. 2017. Omics of bifidobacteria: research and insights into their health-promoting activities. Biochem J 474:4137–4152. doi: 10.1042/BCJ20160756 [DOI] [PubMed] [Google Scholar]
- 18. Tojo R, Suárez A, Clemente MG, de los Reyes-Gavilán CG, Margolles A, Gueimonde M, Ruas-Madiedo P. 2014. Intestinal microbiota in health and disease: role of bifidobacteria in gut homeostasis. World J Gastroenterol 20:15163–15176. doi: 10.3748/wjg.v20.i41.15163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fontana F, Alessandri G, Tarracchini C, Bianchi MG, Rizzo SM, Mancabelli L, Lugli GA, Argentini C, Vergna LM, Anzalone R, Longhi G, Viappiani A, Taurino G, Chiu M, Turroni F, Bussolati O, van Sinderen D, Milani C, Ventura M. 2022. Designation of optimal reference strains representing the infant gut bifidobacterial species through a comprehensive multi-omics approach. Environ Microbiol 24:5825–5839. doi: 10.1111/1462-2920.16205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Turroni F, Milani C, Duranti S, Mahony J, van Sinderen D, Ventura M. 2018. Glycan utilization and cross-feeding activities by bifidobacteria. Trends Microbiol 26:339–350. doi: 10.1016/j.tim.2017.10.001 [DOI] [PubMed] [Google Scholar]
- 21. Turroni F, Milani C, Duranti S, Ferrario C, Lugli GA, Mancabelli L, van Sinderen D, Ventura M. 2018. Bifidobacteria and the infant gut: an example of co-evolution and natural selection. Cell Mol Life Sci 75:103–118. doi: 10.1007/s00018-017-2672-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kelly SM, Munoz-Munoz J, van Sinderen D. 2021. Plant glycan metabolism by bifidobacteria. Front Microbiol 12:609418. doi: 10.3389/fmicb.2021.609418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomson P, Medina DA, Garrido D. 2018. Human milk oligosaccharides and infant gut bifidobacteria: molecular strategies for their utilization. Food Microbiol 75:37–46. doi: 10.1016/j.fm.2017.09.001 [DOI] [PubMed] [Google Scholar]
- 24. Ventura M, O’Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, O’Toole PW. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol 7:61–71. doi: 10.1038/nrmicro2047 [DOI] [PubMed] [Google Scholar]
- 25. Choi IY, Kim J, Kim SH, Ban OH, Yang J, Park MK. 2021. Safety evaluation of Bifidobacterium breve IDCC4401 isolated from infant feces for use as a commercial probiotic. J Microbiol Biotechnol 31:949–955. doi: 10.4014/jmb.2103.03041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ventura M, Turroni F, van Sinderen D. 2012. Probiogenomics as a tool to obtain genetic insights into adaptation of probiotic bacteria to the human gut. Bioeng Bugs 3:73–79. doi: 10.4161/bbug.18540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alessandri G, Fontana F, Tarracchini C, Rizzo SM, Bianchi MG, Taurino G, Chiu M, Lugli GA, Mancabelli L, Argentini C, Longhi G, Anzalone R, Viappiani A, Milani C, Turroni F, Bussolati O, Sinderen D, Ventura M. 2023. Identification of a prototype human gut Bifidobacterium longum Subsp longum strain based on comparative and functional genomic approaches. Front Microbiol 14. doi: 10.3389/fmicb.2023.1130592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berkhout M, Zoetendal E, Plugge C, Belzer C. 2022. Use of synthetic communities to study microbial ecology of the gut. MRR 1:4. doi: 10.20517/mrr.2021.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fernandez-Julia P, Commane DM, van Sinderen D, Munoz-Munoz J. 2022. Cross-feeding interactions between human gut commensals belonging to the bacteroides and Bifidobacterium genera when grown on dietary glycans. MRR 1:12. doi: 10.20517/mrr.2021.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Duranti S, Turroni F, Milani C, Foroni E, Bottacini F, Dal Bello F, Ferrarini A, Delledonne M, van Sinderen D, Ventura M. 2013. Exploration of the genomic diversity and core genome of the Bifidobacterium adolescentis phylogenetic group by means of a polyphasic approach. Appl Environ Microbiol 79:336–346. doi: 10.1128/AEM.02467-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duranti S, Milani C, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Sánchez B, Margolles A, van Sinderen D, Ventura M. 2016. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci Rep 6:23971. doi: 10.1038/srep23971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lugli GA, Duranti S, Albert K, Mancabelli L, Napoli S, Viappiani A, Anzalone R, Longhi G, Milani C, Turroni F, Alessandri G, Sela DA, van Sinderen D, Ventura M. 2019. Unveiling genomic diversity among members of the species Bifidobacterium pseudolongum, a widely distributed gut commensal of the animal kingdom. Appl Environ Microbiol 85:e03065-18. doi: 10.1128/AEM.03065-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lugli GA, Tarracchini C, Alessandri G, Milani C, Mancabelli L, Turroni F, Neuzil-Bunesova V, Ruiz L, Margolles A, Ventura M. 2020. Decoding the genomic variability among members of the Bifidobacterium dentium species. Microorganisms 8:1720. doi: 10.3390/microorganisms8111720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lugli GA, Milani C, Duranti S, Mancabelli L, Mangifesta M, Turroni F, Viappiani A, van Sinderen D, Ventura M. 2018. Tracking the taxonomy of the genus Bifidobacterium based on a phylogenomic approach. Appl Environ Microbiol 84:e02249-17. doi: 10.1128/AEM.02249-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drula E, Garron ML, Dogan S, Lombard V, Henrissat B, Terrapon N. 2022. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res 50:D571–D577. doi: 10.1093/nar/gkab1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Turroni F, Bottacini F, Foroni E, Mulder I, Kim J-H, Zomer A, Sánchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A 107:19514–19519. doi: 10.1073/pnas.1011100107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruas-Madiedo P, Gueimonde M, Fernández-García M, de los Reyes-Gavilán CG, Margolles A. 2008. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol 74:1936–1940. doi: 10.1128/AEM.02509-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bottacini F, Ventura M, van Sinderen D, O’Connell Motherway M. 2014. Diversity, ecology and intestinal function of bifidobacteria. Microb Cell Fact 13 Suppl 1:S4. doi: 10.1186/1475-2859-13-S1-S4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yasmin I, Saeed M, Khan WA, Khaliq A, Chughtai MFJ, Iqbal R, Tehseen S, Naz S, Liaqat A, Mehmood T, Ahsan S, Tanweer S. 2020. In vitro probiotic potential and safety evaluation (hemolytic, cytotoxic activity) of Bifidobacterium strains isolated from raw camel milk. Microorganisms 8:354. doi: 10.3390/microorganisms8030354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Serafini F, Strati F, Ruas-Madiedo P, Turroni F, Foroni E, Duranti S, Milano F, Perotti A, Viappiani A, Guglielmetti S, Buschini A, Margolles A, van Sinderen D, Ventura M. 2013. Evaluation of adhesion properties and antibacterial activities of the infant gut commensal Bifidobacterium bifidum PRL2010. Anaerobe 21:9–17. doi: 10.1016/j.anaerobe.2013.03.003 [DOI] [PubMed] [Google Scholar]
- 41. Russell DA, Ross RP, Fitzgerald GF, Stanton C. 2011. Metabolic activities and probiotic potential of bifidobacteria. Int J Food Microbiol 149:88–105. doi: 10.1016/j.ijfoodmicro.2011.06.003 [DOI] [PubMed] [Google Scholar]
- 42. Achi SC, Halami PM. 2019. In vitro comparative analysis of probiotic and functional attributes of indigenous isolates of bifidobacteria. Curr Microbiol 76:304–311. doi: 10.1007/s00284-018-1615-9 [DOI] [PubMed] [Google Scholar]
- 43. Ruiz L, Ruas-Madiedo P, Gueimonde M, de Los Reyes-Gavilán CG, Margolles A, Sánchez B. 2011. How do bifidobacteria counteract environmental challenges? Mechanisms involved and physiological consequences. Genes Nutr 6:307–318. doi: 10.1007/s12263-010-0207-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Somashekaraiah R, Shruthi B, Deepthi BV, Sreenivasa MY. 2019. Probiotic properties of lactic acid bacteria isolated from neera: a naturally fermenting coconut palm nectar. Front Microbiol 10:1382. doi: 10.3389/fmicb.2019.01382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Turroni F, Serafini F, Foroni E, Duranti S, O’Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sánchez B, et al. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum Prl2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A 110:11151–11156. doi: 10.1073/pnas.1303897110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rizzo SM, Alessandri G, Lugli GA, Fontana F, Tarracchini C, Mancabelli L, Viappiani A, Bianchi MG, Bussolati O, van Sinderen D, Ventura M, Turroni F. 2023. Exploring molecular interactions between human milk hormone insulin and bifidobacteria. Microbiol Spectr 11:e0066523. doi: 10.1128/spectrum.00665-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guglielmetti S, Tamagnini I, Mora D, Minuzzo M, Scarafoni A, Arioli S, Hellman J, Karp M, Parini C. 2008. Implication of an outer surface lipoprotein in adhesion of Bifidobacterium bifidum to caco-2 cells. Appl Environ Microbiol 74:4695–4702. doi: 10.1128/AEM.00124-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Valeriano VD, Parungao-Balolong MM, Kang DK. 2014. In vitro evaluation of the mucin-adhesion ability and probiotic potential of Lactobacillus mucosae LM1. J Appl Microbiol 117:485–497. doi: 10.1111/jam.12539 [DOI] [PubMed] [Google Scholar]
- 49. Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. 2016. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol 7:979. doi: 10.3389/fmicb.2016.00979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG. 2015. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett 362:fnv176. doi: 10.1093/femsle/fnv176 [DOI] [PubMed] [Google Scholar]
- 51. Saedisomeolia A, Wood LG, Garg ML, Gibson PG, Wark PAB.. 2009. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101:533–540. [DOI] [PubMed] [Google Scholar]
- 52. Turroni F, Serafini F, Mangifesta M, Arioli S, Mora D, van Sinderen D, Ventura M. 2014. Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol Lett 357:23–33. doi: 10.1111/1574-6968.12509 [DOI] [PubMed] [Google Scholar]
- 53. Bunesova V, Lacroix C, Schwab C. 2018. Mucin cross-feeding of infant bifidobacteria and Eubacterium hallii. Microb Ecol 75:228–238. doi: 10.1007/s00248-017-1037-4 [DOI] [PubMed] [Google Scholar]
- 54. Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, Li J, Palanza P, Delledonne M, van Sinderen D, Ventura M. 2016. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J 10:1656–1668. doi: 10.1038/ismej.2015.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y. 2005. Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl Environ Microbiol 71:7099–7106. doi: 10.1128/AEM.71.11.7099-7106.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fondevila M, Dehority BA. 1994. Degradation and utilization of forage hemicellulose by rumen bacteria, singly in coculture or added sequentially. J Appl Bacteriol 77:541–548. doi: 10.1111/j.1365-2672.1994.tb04399.x [DOI] [PubMed] [Google Scholar]
- 57. Mihăşan M, Brandsch R. 2016. A predicted T4 secretion system and conserved DNA-repeats identified in a subset of related Arthrobacter plasmids. Microbiol Res 191:32–37. doi: 10.1016/j.micres.2016.05.008 [DOI] [PubMed] [Google Scholar]
- 58. Aw W, Fukuda S. 2019. Protective effects of bifidobacteria against enteropathogens. Microb Biotechnol 12:1097–1100. doi: 10.1111/1751-7915.13460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ruiz L, Delgado S, Ruas-Madiedo P, Sánchez B, Margolles A. 2017. Bifidobacteria and their molecular communication with the immune system. Front Microbiol 8. doi: 10.3389/fmicb.2017.02345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O’Toole PW, van Sinderen D, Marchesi JR, Ventura M, Neu J. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 7:e36957. doi: 10.1371/journal.pone.0036957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Milani C, Lugli GA, Fontana F, Mancabelli L, Alessandri G, Longhi G, Anzalone R, Viappiani A, Turroni F, van Sinderen D, Ventura M, Arumugam M. 2021. METannotatorx2: a comprehensive tool for deep and shallow metagenomic data set analyses. mSystems 6:mSystems doi: 10.1128/mSystems.00583-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chen Y, Ye W, Zhang Y, Xu Y. 2015. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res 43:7762–7768. doi: 10.1093/nar/gkv784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Olm MR, Brown CT, Brooks B, Banfield JF. 2017. dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J 11:2864–2868. doi: 10.1038/ismej.2017.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27:722–736. doi: 10.1101/gr.215087.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wick RR, Holt KE. 2022. Polypolish: short-read polishing of long-read bacterial genome assemblies. PLoS Comput Biol 18:e1009802. doi: 10.1371/journal.pcbi.1009802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lugli GA, Fontana F, Tarracchini C, Milani C, Mancabelli L, Turroni F, Ventura M. 2023. MEGAnnotator2: a pipeline for the assembly and annotation of microbial genomes. Microbiome Res Rep 2:15. doi: 10.20517/mrr.2022.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform 11:119. doi: 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. 2016. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol Lett 363:fnw049. doi: 10.1093/femsle/fnw049 [DOI] [PubMed] [Google Scholar]
- 71. Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinform 28:416–418. doi: 10.1093/bioinformatics/btr655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. doi: 10.1093/nar/gky418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mancabelli L, Mancino W, Lugli GA, Argentini C, Longhi G, Milani C, Viappiani A, Anzalone R, Bernasconi S, van Sinderen D, Ventura M, Turroni F. 2021. Amoxicillin-clavulanic acid resistance in the genus Bifidobacterium. Appl Environ Microbiol 87:e03137-20. doi: 10.1128/AEM.03137-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dostal A, Lacroix C, Bircher L, Pham VT, Follador R, Zimmermann MB, Chassard C, Huffnagle GB. 2015. Iron modulates butyrate production by a child gut microbiota in vitro. mBio 6:mBio. doi: 10.1128/mBio.01453-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wylensek D, Hitch TCA, Riedel T, Afrizal A, Kumar N, Wortmann E, Liu T, Devendran S, Lesker TR, Hernández SB, et al. 2020. A collection of bacterial isolates from the pig intestine reveals functional and taxonomic diversity. Nat Commun 11. doi: 10.1038/s41467-020-19929-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Aranda-Díaz A, Ng KM, Thomsen T, Real-Ramírez I, Dahan D, Dittmar S, Gonzalez CG, Chavez T, Vasquez KS, Nguyen TH, Yu FB, Higginbottom SK, Neff NF, Elias JE, Sonnenburg JL, Huang KC. 2022. Establishment and characterization of stable, diverse, fecal-derived in vitro microbial communities that model the intestinal microbiota. Cell Host Microbe 30:260–272. doi: 10.1016/j.chom.2021.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Schwab C, Ruscheweyh HJ, Bunesova V, Pham VT, Beerenwinkel N, Lacroix C. 2017. Trophic interactions of infant bifidobacteria and Eubacterium hallii during L-fucose and fucosyllactose degradation. Front. Microbiol 8. doi: 10.3389/fmicb.2017.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Scott KP, Martin JC, Duncan SH, Flint HJ. 2014. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol Ecol 87:30–40. doi: 10.1111/1574-6941.12186 [DOI] [PubMed] [Google Scholar]
- 79. Henriques A, Cereija T, Machado A, Cerca N. 2012. In silico vs in vitro analysis of primer specificity for the detection of Gardnerella vaginalis, Atopobium vaginae and Lactobacillus spp . BMC Res Notes 5. doi: 10.1186/1756-0500-5-637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Milani C, Lugli GA, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, Arioli S, Sanchez B, Lane J, Ward DV, Hickey R, Mora D, Segata N, Margolles A, van Sinderen D, Ventura M. 2015. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep 5. doi: 10.1038/srep15782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Milani C, Alessandri G, Mancabelli L, Mangifesta M, Lugli GA, Viappiani A, Longhi G, Anzalone R, Duranti S, Turroni F, Ossiprandi MC, van Sinderen D, Ventura M, Björkroth J. 2020. Multi-omics approaches to decipher the impact of diet and host physiology on the mammalian gut microbiome. Appl Environ Microbiol 86. doi: 10.1128/AEM.01864-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Langdon WB. 2015. Performance of genetic programming optimised bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min 8:1. doi: 10.1186/s13040-014-0034-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Anders S, Pyl PT, Huber W. 2015. Htseq--a python framework to work with high-throughput sequencing data. Bioinform 31:166–169. doi: 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data . Bioinform 26:139–140. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Paradis E, Schliep K. 2019. Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinform 35:526–528. doi: 10.1093/bioinformatics/bty633 [DOI] [PubMed] [Google Scholar]
- 86. Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. 2001. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284. doi: 10.1016/s0166-4328(01)00297-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative PCR evaluation of the numerical load of B. adolescentis PRL2023.
Adhesion of B. adolescentis PRL2023 and ATCC 15703 and B. animalis subsp. lactis Bb-12 cells to HT29-X cells monolayers.
Supplemental tables.
Data Availability Statement
Raw sequences of metatranscriptomics experiments were accessible through the SRA study BioProject PRJNA1011847. Furthermore, the updated genome sequence of B. adolescentis PRL2023 was deposited in the GenBank database with the NCBI RefSeq accession code CP133648.