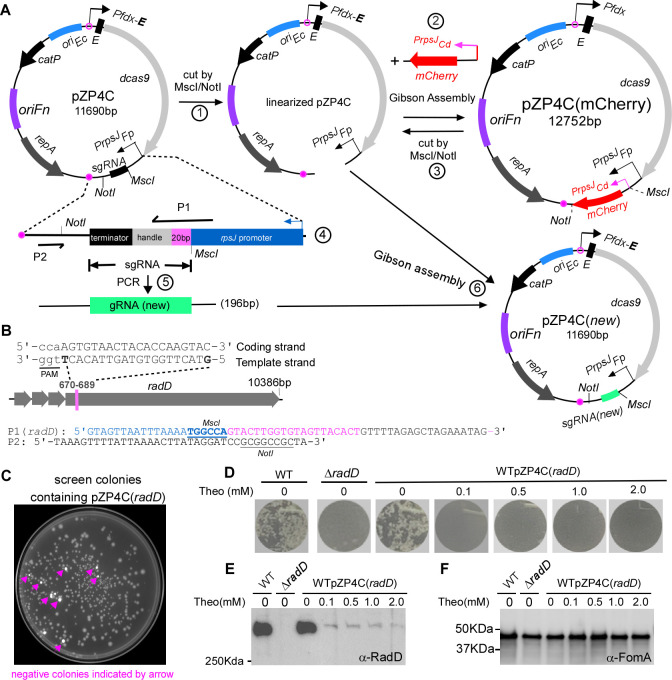
Fig 2.
Use of mCherry as a reporter for streamlined screening of positive CRISPRi clones and exemplifying CRISPRi targeting with radD. (A) Diagram illustrating the construction of CRISPRi plasmid with mCherry as a screening reporter is outlined. (1) Linearization of pZP4C: the pZP4C is cleaved using MscI and NotI enzymes. (2) Replacement of sgRNA segment with the mCherry gene. The mCherry is driven by the PrpsJCd promoter. The resulting plasmid is designated as pZP4C(mCherry). (3) Release of PrpsJCd -mCherry: pZP4C(mCherry) is again cut using MscI/NotI to release PrpsJCd -mCherry for subsequent cloning. (4) Design primers P1 and P2. P1 is a 60-nt primer starting with a 21-nt PrpsJFP promoter match, followed by a 20-nt target gene sequence and ending with a 19-nt sgRNA handle region. The sgRNA includes a 20-nt base-pairing region, a 42-nt dCas9-binding RNA structure (dCas9 handle), and a 40-nt transcription termination sequence from S. pyogenes. (5) Amplification of new target sgRNA: use pZP4C plasmid DNA as a template with primers P1 and P2 to generate a 196-bp target sgRNA fragment. (6) Cloning of new sgRNA: insert the new sgRNA fragment into digested pZP4C(mCherry) via Gibson assembly, creating a new CRISPRi plasmid for targeting a new gene. (B) Designing P1 primer with radD gene example. For instance, the radD gene illustrates the P1 primer design. A 20-nt base pair region from the 670th to the 789th base of radD is selected. This sequence, aligned in a 5′ to 3′ orientation on the template strand, is incorporated into the P1 primer [designated P1(radD)]. The PAM (TGG) sequence is indicated through underlining in lowercase. The MscI site in P1 and the NotI site in P2 are shown. (C) Positive clone selection uses a ChemiDoc MP Imaging System (Bio-Rad). Fluorescent cells, considered negative clones, appear pseudocolored in white under the Cy3 setting, whereas non-fluorescent cells (positive clones) are in gray. (D) CRISPRi-mediated repression of the radD gene abolishes coaggregation with A. oris. The wild-type F. nucleatum strain containing pZP4C(radD) was cultured in a TSPC medium under different theophylline concentrations for 22 hours in anaerobic conditions. Subsequently, cells were collected, washed, resuspended in a coaggregation buffer, and assessed for coaggregation with A. oris. A representative result is presented after three experimental repeats. (E and F) Cells subjected to the experimental conditions detailed in panel D undergo SDS-PAGE followed by immunoblotting. Antibodies specific to RadD (α-RadD) and FomA (α-FomA) are used for detection. FomA serves as a loading control. Molecular weight markers (in kilodaltons) are indicated on the left side of the blot.