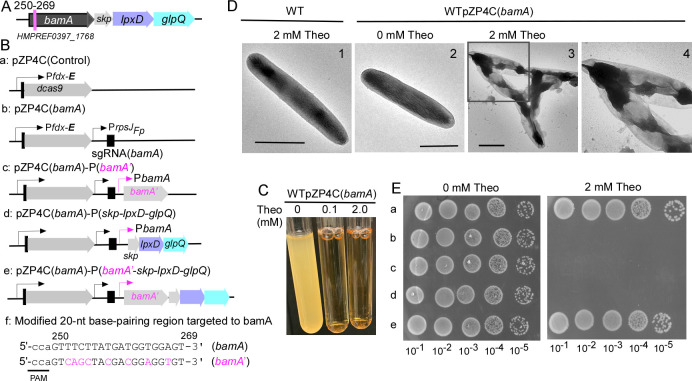
Fig 3.
CRISPRi targeting essential gene bamA. (A) Shown is the bamA gene operon in F. nucleatum. This operon consists of four consecutive genes: bamA, skp, lpxD, and glpQ. The specific 20-nt base-pairing region within the bamA gene, which is targeted, is marked with a red line, along with the locus number for reference. (B) A schematic illustrates various bamA-related CRISPRi plasmids. These include pZP4C(bamA) (b) and a control plasmid (a) containing dCas9 without sgRNA. Additionally, three modified plasmids derived from pZP4C(bamA) are depicted: expressing bamA (c), the triplet genes skp-lpxD-glpQ (d), and the entire four - gene operon (e). Notably, the sequence expressing bamA in c and e has been modified to prevent CRISPRi recognition, with altered nucleotides highlighted in red. The modified bamA is referred to as bamA′. (C) CRISPRi-mediated silencing of bamA leads to growth inhibition. Wild-type cells containing pZP4C(bamA) were cultivated in a TSPC medium supplemented with varying concentrations (0, 0.1, and 2.0 mM) of theophylline for 22 hours. (D) bamA silencing induces short filamentation and alters the outer membrane structure. Cells with pZP4C(bamA) under 0 or 2 mM theophylline were immobilized on carbon-coated nickel grids, stained with 0.1% uranyl acetate, and observed using a transmission electron microscope. Enlarged views of specific areas (D3) are displayed in D4: scale bar, 1 µm. (E) The rescue of the death phenotype caused by CRISPRi-mediated bamA silencing is demonstrated. Cells containing one of the various plasmids shown in panel B were cultured overnight without inducers. Subsequently, these cultures were serially diluted 10-fold and spotted on agar plates with and without inducers. Photographs of the plates were taken after incubation in anaerobic conditions for 3 days.