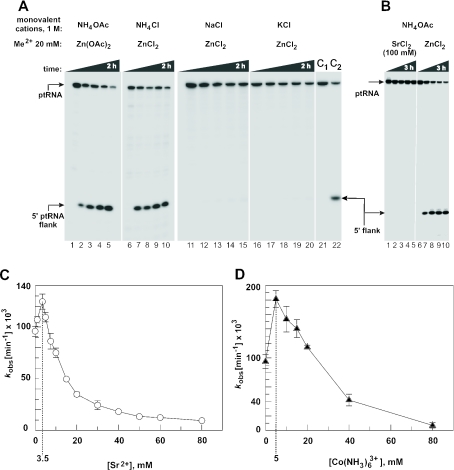
Figure 2.
Processing of the all-ribose ptRNAGly by E.coli M1 RNA under single turnover conditions at 37°C in the presence of 20 mM Zn2+. (A) Dependence on the type of monovalent cation; processing occurred in the presence of ammonium, but not sodium or potassium salts; reaction conditions: 5 μM M1 RNA, <1 nM ptRNA, 50 mM PIPES, pH 6.65, 1 M monovalent salt as indicated, and 20 mM ZnCl2 or Zn(OAc)2; control lanes 1, 6, 11, 16 and 21: incubation for 2 h in the absence of M1 RNA; time points were 5 min (lanes 2, 7, 12, 17 and 22), 20 min (lanes 3, 8, 13 and 18), 1 h (lanes 4, 9, 14 and 19) and 2 h (lanes 5, 10, 15 and 20); lanes 21 and 22 (C1, C2) are equal to lanes 1 and 2. (B) Assay documenting that no processing occurs in the presence of Sr2+ as the sole metal ion; reaction conditions: 5 μM M1 RNA, <1 nM ptRNA, 50 mM PIPES, pH 6.65, 1 M NH4OAc and 100 mM SrCl2 (left) or 20 mM ZnCl2 (right). Control lanes 1 and 6: incubation for 3 h in the absence of M1 RNA; time points were 5 min (lane 7), 20 min (lanes 2 and 8), 40 min (lane 3), 1 h (lanes 4 and 9) and 3 h (lanes 5 and 10). (C and D) Processing rates as a function of increasing concentrations of SrCl2 (C) or Co(NH3)6Cl3 (D); reaction conditions: 5 μM M1 RNA, <1 nM ptRNA, 50 mM PIPES, pH 6.65, 1 M NH4OAc and 20 mM Zn(OAc)2; transition points between the stimulatory and the inhibitory phases of the curves are marked by dashed lines.