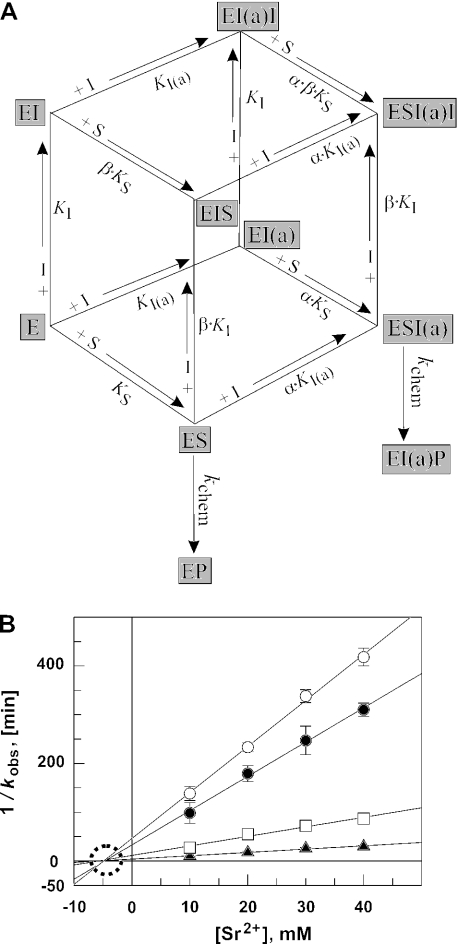
Figure 8.
(A) Equilibria for the M1 RNA-catalyzed processing reaction in the presence of constant [Zn2+] as the catalytic cofactor and varying concentrations of [Sr2+] that activate the reaction at low but inhibit at higher concentrations. The model involves two Sr2+ ions (or two classes of Sr2+ ions) that both improve substrate affinity in a cooperative manner (see Figure 3B), but one of the two inhibiting substrate conversion non-competitively with respect to the substrate. Other models, for example, assuming the involvement of two ‘activating’ and one inhibitory Sr2+ ion or predicting that E·S·I and E·S·I(a)·I complexes retain residual reactivity, failed to give satisfactory curve fits of the data in Figure 7C and D. I(a) = Sr2+ ion that activates processing by decreasing KS to αKS; I = inhibitory Sr2+ ion that inhibits cleavage chemistry but also decreases KS to βKS. (B) Dixon plot of the M1 RNA-catalyzed ptRNA cleavage rate at 37°C as a function of [Sr2+] in the presence of four different fixed Zn2+ concentrations and 5 μM M1 RNA, <1 nM ptRNA, 50 mM PIPES, pH 6.65 and 1 M NH4OAc; open circles, 8.1 mM Zn2+; filled circles, 10 mM Zn2+; open squares, 15 mM Zn2+; and filled triangles, 20 mM Zn2+. All four datasets fit to straight lines intersecting on the x-axis, indicating that Sr2+ acts as a non-competitive inhibitor with respect to Zn2+. The point of intersection on the x-axis yields a KI-value of ∼5 mM.