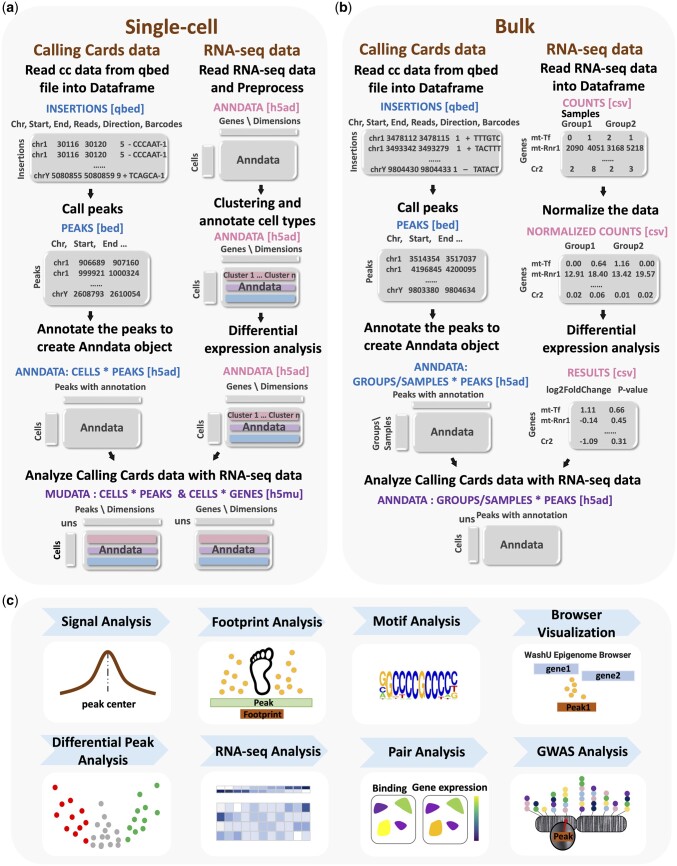
Figure 1.
Overview of Pycallingcards. (a) Pycallingcards workflow for scCC data. Pycallingcards reads insertion data from a qbed file and then calls peaks (to create a bed file, left column). It then creates a cells-by-peaks Anndata object (h5ad file) Pycallingcards interfaces with Scanpy to complete preprocessing, clustering, and differential expression analysis of the RNA-seq data collected for each cell (right column). Pycallingcards then uses Mudata object to store the combined scCC and scRNA-seq data (h5mu file). (b) Data structure in Pycallingcards for bulk CC data. Pycallingcards reads insertion data from a qbed file and calls peaks, which generates a bed file. It later creates a groups/samples-by-peaks Anndata object (h5ad file) (b, left column). If bulk RNA-seq is provided, it uses normalized counts and results from differential gene analysis (b, right column). (c) Downstream Analysis. Pycallingcards provides functionality to compare called CC peaks with Chip-seq signal (when available), perform a footprint analysis to narrow down TF binding regions, find motifs, allow for visualization of the dataset through the WashU Epigenome Browser, perform differential peak analysis, pair CC data with RNA-seq data, and identify related SNPs by intersecting peaks with a GWAS database.