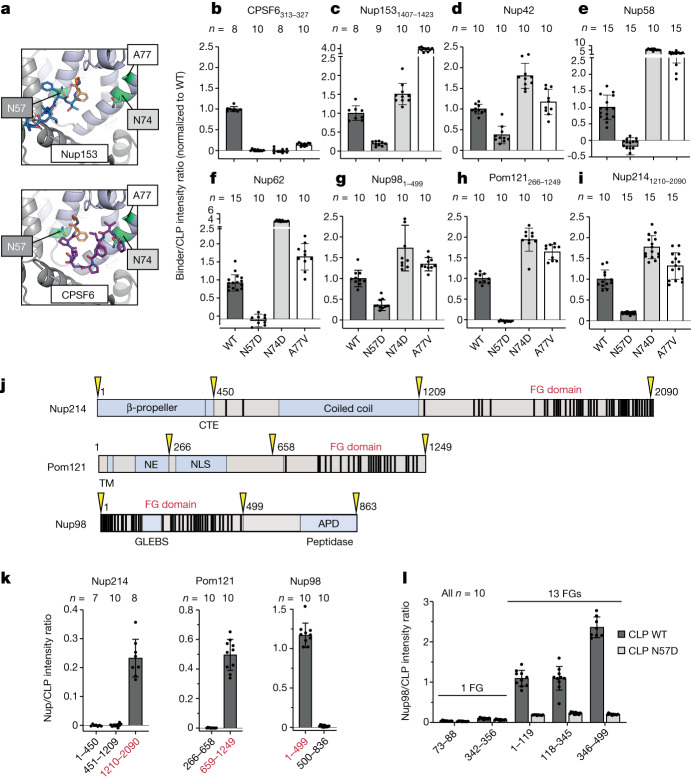
Fig. 2. HIV-1 CA specifically recruits FG-repeat domains.
a, Hydrophobic pocket of CA (cartoon) with Nup153 (top, blue; PDB 4U0C) and CPSF6 (bottom, purple; PDB 4U0B) peptides bound. Buried FG motifs are shown in orange, with key CA residues in green. b–i, Binding of CLPs (wild-type or mutant) to CPSF6313–327 (b), Nup1531407–1423 (c), Nup42 (d), Nup58 (e), Nup62 (f), Nup981–499 (g), Pom121266–1249 (h) or Nup2141210–2090 (i), as measured by FFS. j, Domain architectures of Nup214, Pom121 and Nup98. FG motifs are shown as black bars. CTE, C-terminally extended peptide; TM, transmembrane domain; NE, inner nuclear envelope-binding region; NLS, nuclear localization signal; GLEBS, Gle2-binding sequence; APD, autoproteolytic domain. Domain boundaries of truncation constructs are shown in yellow. k, Binding of truncated constructs of Nup214, Pom121 and Nup98 to CLPs as measured by FFS. FG-repeat domains are shown in red. l, Binding of Nup98 peptides and fragments of Nup98 to CLPs. Error bars show standard deviation of the mean; n, number of scans per sample.