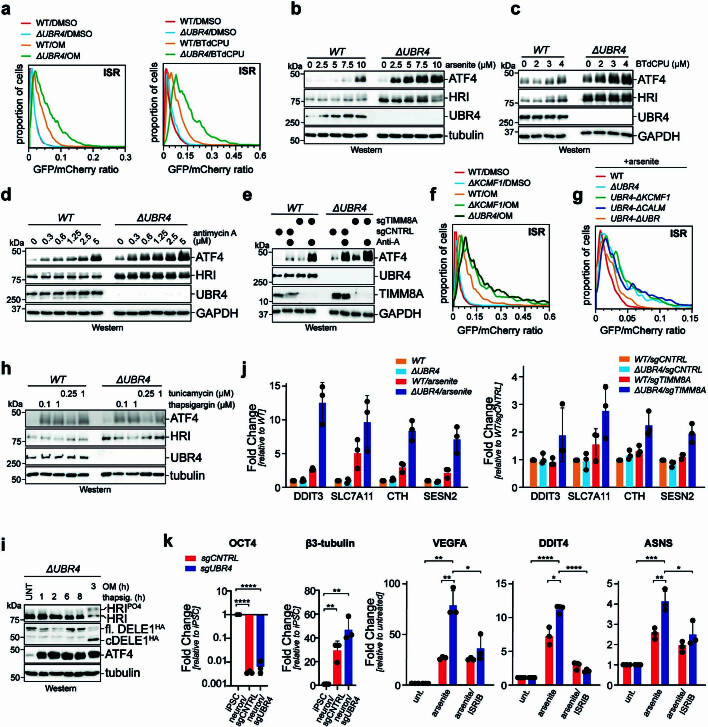
Extended Data Fig. 5. The SIFI complex silences the integrated stress response.
a. Deletion of UBR4 increases ISR signaling in response to cells being treated with oligomycin (0.2 μM) for 16 h or BTdCPU (7.5 μM) for 8 h. ISR activation was monitored by flow cytometry using the uORF-ATF4 reporter described above. Similar results in n = 2 independent experiments. b. UBR4 deletion increases ISR signaling. Wildtype or ΔUBR4 cells were treated for 16 h with increasing concentrations of arsenite and analyzed for ATF4 levels by Western blotting. Similar results in n = 2 independent experiments. c. Deletion of UBR4 increases ISR signaling in cells treated for 16 h with increasing concentrations of BTdCPU, as monitored by Western blots detecting ATF4. Similar results in n = 2 independent experiments. d. UBR4 deletion increases ISR signaling in cells treated for 16 h with increasing concentrations of antimycin A, as detected by ATF4 expression. Similar results in n = 2 independent experiments. e. Deletion of TIMM8A induces ATF4 accumulation more strongly in ΔUBR4 cells. WT or ΔUBR4 cells depleted of TIMM8A were treated with antimycin A (0.6 μM) for 16 h. Similar results in n = 2 independent experiments. f. Deletion of KCMF1 increases ISR signaling to a similar extent as UBR4 deletion, as detected using the uORF-dependent ISR reporter in flow cytometry. Cells were treated with OM (0.2 μM) for 8 h. Similar results in n = 2 independent experiments g. Deletion of KCMF1- and calmodulin-binding domains in the endogenous UBR4 locus increases ISR signaling in response to 5 μM sodium arsenite for 16 h, as determined by flow cytometry using the uORF-dependent ISR reporter. Similar results in n = 2 independent experiments h. UBR4 does not restrict ISR signaling in response to endoplasmic reticulum stress. Wildtype or ΔUBR4 cells were treated with thapsigargin or tunicamycin for 8 h and analyzed for ATF4 levels by Western blotting. Experiment performed once. i. ER stress activation by thapsigargin does not induce DELE1 cleavage. Cells were treated with thapsigargin (1 μM) or oligomycin (1 μM) for the indicated times. Experiment performed once. j. UBR4 deletion increases ISR signaling, as read out by ATF4 activation. Wildtype or ΔUBR4 cells were either treated with 5 μM sodium arsenite (left panel) or depleted of TIMM8A (right panel) and expression of established ATF4 target genes was determined by qPCR. Graph shows mean ± SD of 3 independent experiments. k. UBR4 depletion increases ISR signaling in neurons derived from induced pluripotent stem cells by NGN2 activation. Differentiation was ensured by qRT-PCR against OCT4 and β3-tubulin and ISR target gene expression was measured by qRT-PCR. As indicated, either 5 μM sodium arsenite or ISRIB were added. Graph shows mean ± SD of 3 independent experiments. Statistical significance was determined using a two-tailed Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001. Exact p-Values: OCT4: sgCNTRL p < 0.0001; sgUBR4 p < 0.0001. β3-tubulin: sgCNTRL p = 0.0046; sgUBR4 p = 0.0014. VEGFA: sgUBR4 arsenite vs. unt. p = 0.0011; sgUBR4 arsenite vs. sgCNTRL arsenite p = 0.0052; sgUBR4 arsenite/ISRIB vs. sgUBR4 arsenite p = 0.0242. DDIT4: sgUBR4 arsenite vs. unt. p < 0.0001; sgUBR4 arsenite vs. sgCNTRL arsenite p = 0.0143; sgUBR4 arsenite/ISRIB vs. sgUBR4 arsenite p < 0.0001. ASNS: sgUBR4 arsenite vs. unt. p = 0.0004; sgUBR4 arsenite vs. sgCNTRL arsenite p = 0.0089; sgUBR4 arsenite/ISRIB vs. sgUBR4 arsenite p = 0.0236. For gel source data, see Supplementary Fig. 1.