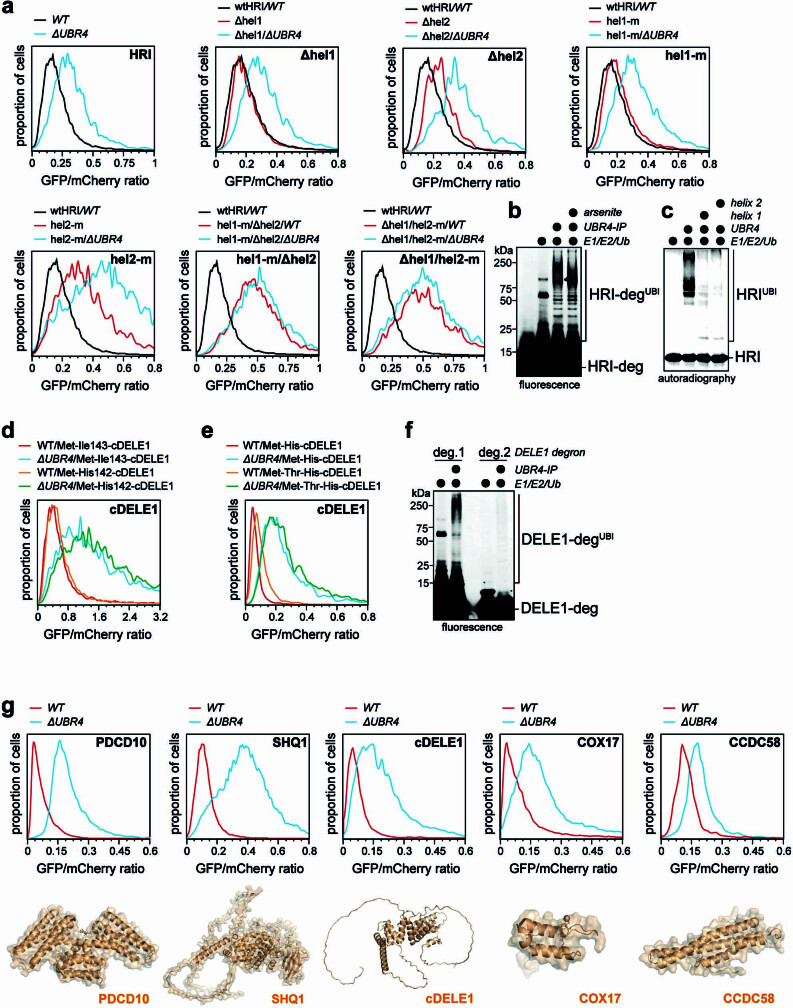
Extended Data Fig. 7. The SIFI complex detects helical degrons in HRI and DELE1.
a. Deletion or mutation of two helices in HRI at the same time, but not manipulation of a single helix, protects HRI from UBR4-dependent degradation. The stability of indicated mutants was analyzed in wildtype or ΔUBR4 cells by flow cytometry using the GFP/mCherry-based degradation reporter. Similar results in n ≥ 2 independent experiments. b. The SIFI complex ubiquitylates a single HRI peptide irrespectively of whether the SIFI complex was purified from control cells or cells treated with arsenite (40 μM for 4 h). Experiment performed once. c. Peptides encompassing a single HRI helix compete for ubiquitylation of the entire amino-terminal HRI domain (residues 1–138). 35S-labeled HRI1–138-SUMO was incubated with affinity-purified SIFI complexes, E1, UBE2A and UBE2D3, and ubiquitin. 200 μM of purified peptides encompassing the helices comprising degron 1 or degron 2, respectively, were added, and reaction products were analyzed by autoradiography. Similar results in n = 2 independent experiments d. Changing the amino-terminus of cleaved DELE1 does not affect its stability, as seen by flow cytometry. Similar results in n = 2 independent experiments e. Capping of the amino-terminus of cleaved DELE1 with threonine, an amino acid not recognized by the N-end rule, does not change its stability, as seen by flow cytometry. Similar results in n = 2 independent experiments. f. A helical DELE1 degron similar to HRI helices is ubiquitylated by the SIFI complex as a TAMRA-labeled peptide, while a distinct DELE1 peptide was not modified. Similar results in n = 2 independent experiments. g. Other top SIFI substrates are mostly composed of α-helices. The stability of top SIFI substrates identified in our screen was analyzed in wildtype or ΔUBR4 cells by flow cytometry, using our degradation reporters. Similar results in n ≥ 2 independent experiments. AlphaFold2 models of each substrate are shown below. For gel source data, see Supplementary Fig. 1.