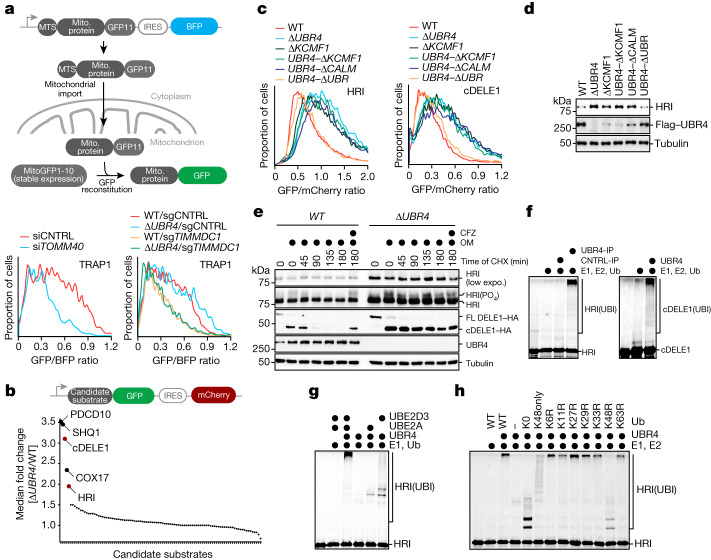
Fig. 2. SIFI targets DELE1 and HRI.
a, Top, outline of the import assay. Bottom, import assay using the model protein TRAP1 in WT or ΔUBR4 cells lacking TOMM40 (left) or the UBR4 genetic interactor TIMMDC1 (right). Similar results in n = 3 independent experiments. Mito., mitochondrial; si, small interfering RNA b, Stability reporter-based screen for UBR4 substrates identifies cleaved DELE1 and HRI. Upper schematic: map of reporter construct with GFP-tagged candidate substrate co-expressed with mCherry under control of an internal ribosome entry site (IRES). c, cDELE1 or HRI stability were monitored by flow cytometry (UBR4–ΔKCMF1: KCMF1-binding domain deleted in endogenous UBR4; same for the other domains). Similar results in n = 2 independent experiments. d, Endogenous HRI increases in cells lacking SIFI, as measured by western blotting. Similar results in n = 3 independent experiments. e, WT or ΔUBR4 cells that expressed endogenously tagged DELE1–HA were exposed to oligomycin (OM, 1 μM) before cycloheximide (CHX) and analysed by western blotting. Where indicated, carfilzomib (CFZ) was added. Similar results in n = 2 independent experiments. expo., exposure; FL, full length. f, 35S-labelled cDELE1(142–515)–SUMO or HRI(1–138)–SUMO were ubiquitylated by SIFI, E1, UBE2A, UBE2D3 and ubiquitin (Ub). Similar results in n = 3 independent experiments. IP, immunoprecipitation. g, SIFI-dependent ubiquitylation requires UBE2A and UBE2D3. Similar results in n = 2 independent experiments. h, SIFI assembles predominantly K48-linked ubiquitin chains. (K0, all Lys mutated; K48only, only Lys48 present; K48R, only Lys48 mutated). Similar results in n = 2 independent experiments. For gel source data, see Supplementary Fig. 1.