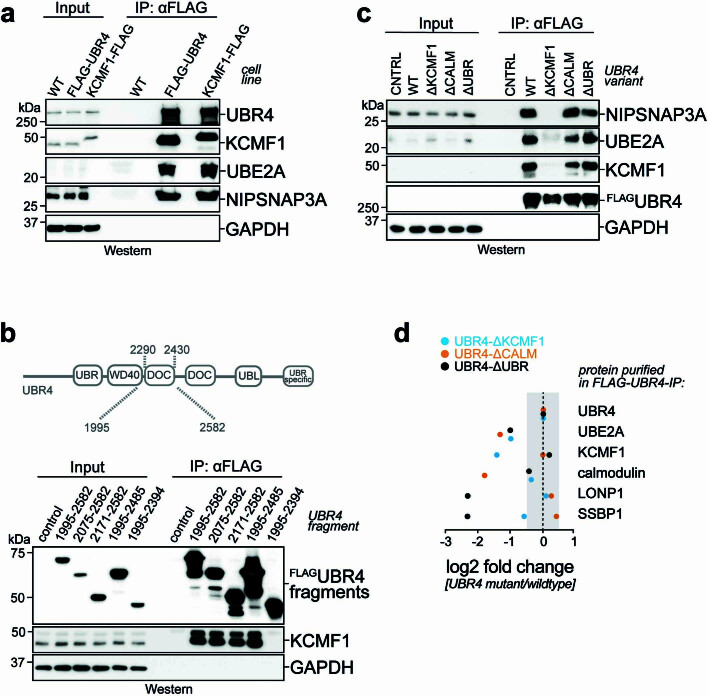
Extended Data Fig. 2. UBR4 stably interacts with KCMF1.
a. Endogenously FLAG-tagged UBR4 and KCMF1 were affinity-purified from 293T cells, and bound proteins were detected by Western blotting using specific antibodies. Similar results in n = 3 independent experiments. b. KCMF1 binds to a DOC domain in UBR4. FLAG-tagged fragments of UBR4 were immunoprecipitated from ΔUBR4 cells and co-precipitating endogenous KCMF1 was detected by Western blotting. Similar results in n = 2 independent experiments. c. Validation of the KCMF1 domain in endogenous UBR4. The DOC domain of UBR4 was excised from the endogenous UBR4 locus (UBR4 was already fused to a FLAG epitope) by CRISPR-Cas9 genome engineering. Similar approaches were used to eliminate the endogenous UBR- and calmodulin-binding regions in UBR4. Endogenous wildtype or mutant UBR4 was affinity-purified, and co-precipitating proteins were detected by Western blotting. Similar results in n = 3 independent experiments. d. Validation of mutant UBR4 by mass spectrometry. The KCMF1- or calmodulin-binding domains, or the UBR domain, were deleted in the endogenous locus of FLAGUBR4. Endogenous UBR4 complexes were affinity-purified and bound proteins were detected by mass spectrometry. Changes in interactions for mutant cell lines compared to wildtype UBR4 are depicted for select proteins. Spectral counts were normalized to bait (UBR4) spectral counts in each cell line. For gel source data, see Supplementary Fig. 1.