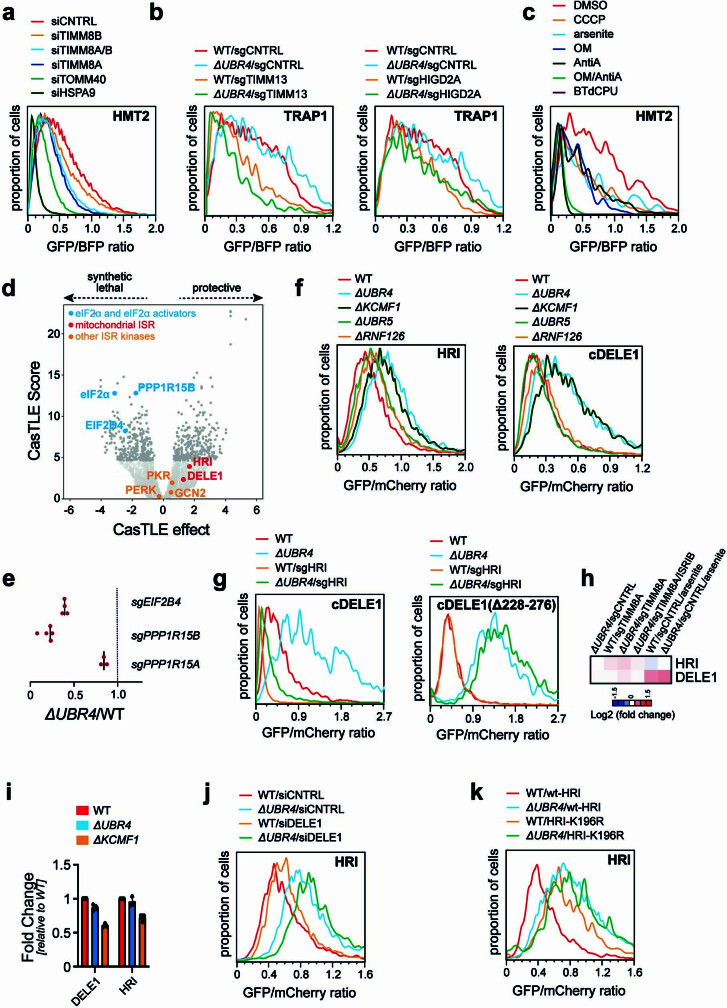
Extended Data Fig. 3. The SIFI complex targets cleaved DELE1 and HRI.
a. Genetic interactors of UBR4 control mitochondrial protein import. Mitochondrial import of GFP11-tagged HMT2 was monitored in WT cells stably expressing mitochondrially targeted GFP(1–10). Genetic interactors of UBR4 or known protein import regulators were depleted with specific siRNAs. Similar results in n = 2 independent experiments. b. UBR4 does not regulate mitochondrial protein import. Import of GFP11-tagged TRAP1 was analyzed as above. When indicated, ΔUBR4 cells were used or the genetic interactors of UBR4, TIMM13 and HIGD2A, were depleted using sgRNAs. Similar results in n = 3 independent experiments. c. Chemical stressors that deplete ΔUBR4 cells in competition assays, compromised mitochondrial protein import. Import of GFP11-tagged HMT2 was analyzed in the presence of indicated drugs CCCP(5 μM), arsenite (10 μM), OM (2.5 μM), Antimycin A (10 μM), BTdCPU (10 μM) for 16 h by flow cytometry, as described above. Similar results in n = 2 independent experiments. d. Depletion of eIF2α, the eIF2B subunit EIF2B4, or the eIF2α phosphatase PPP1R15B causes synthetic lethality with loss of UBR4, as seen in our synthetic lethality screen described earlier. e. Validation of synthetic lethality between EIF2B4 and PPP1R15B (CReP) by cell competition assays. The second eIF2α phosphatase PPP1R15A (GADD34) also shows weak synthetic lethality with UBR4 deletion. f. HRI and cleaved DELE1 are degraded through UBR4 and KCMF1, while the quality control E3 ligases UBR5 or RNF126 are not required. E3 ligases were deleted from 293 T cells by CRISPR/Cas9-mediated genome engineering and the stability of HRI or cDELE1 was monitored as GFP-tagged proteins by flow cytometry. Experiment performed once, similar results obtained with siRNA depletions. g. Degradation of orphan cDELE1, which is not bound to HRI, requires a central domain in cDELE1. A stability reporter expressing either cDELE1 or an internal deletion resistant to HRI depletion (cDELE1Δ228–276) were monitored by flow cytometry in either wildtype or ΔUBR4 cells. When indicated, HRI was depleted by specific sgRNAs. Similar results in n = 2 independent experiments. h. Expression of HRI or DELE1 is not induced by mitochondrial stress, as seen by RNAseq in cells depleted of TIMM8A or treated with arsenite. Data was taken from RNAseq experiments described in Fig. 3d. i. Expression of HRI or DELE1 is not induced by deletion of UBR4 or KCMF1, as seen by qRT-PCR. Graph shows mean ± SD of 3 independent experiments. j. Degradation of an overexpressed wildtype HRI reporter does not require DELE1. Stability of the HRI reporter was monitored in cells treated with control siRNAs or siRNAs targeting DELE1, by flow cytometry. Experiment performed once. k. Mutation of K196 in HRI, which is required for autophosphorylation and activation, prevents UBR4-dependent degradation, as seen by flow cytometry. Similar results in n = 3 independent experiments.