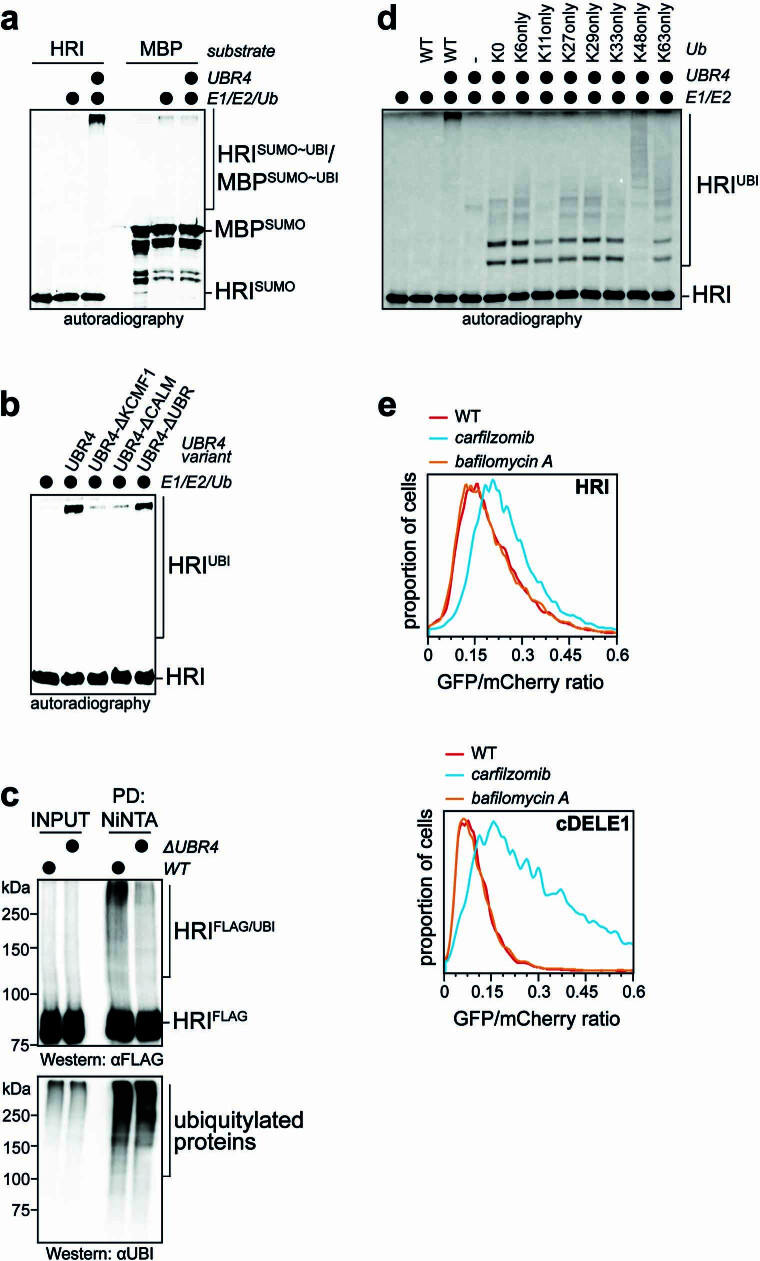
Extended Data Fig. 4. The SIFI complex targets HRI and cDELE1 for proteasomal degradation.
a. HRI is ubiquitylated by the SIFI complex. The SIFI complex was purified from 293 T cells expressing endogenously FLAG-tagged UBR4. It was incubated with 35S-labeled HRI1–138-SUMO or MBP-SUMO as a control, respectively, E1, UBE2D3 and UBE2A as E2 enzymes, and ubiquitin. Reaction products were visualized by autoradiography. Experiment performed once. b. All SIFI subunits are required for HRI ubiquitylation. SIFI complexes were purified from cells expressing endogenously FLAG-tagged UBR4. When indicated, cells with internal deletions of the KCMF1-binding domain, the calmodulin-binding domain, or the UBR domain in endogenous UBR4 were used. 35S-labeled HRI1–138-SUMO, E1, UBE2D3, UBE2A and ubiquitin were added, and reaction products were visualized by autoradiography. Similar results in n = 2 independent experiments. c. The SIFI complex mediates HRI ubiquitylation in cells. Ubiquitin conjugates were purified under denaturing conditions from cells expressing HRIFLAG and HISubiquitin, and modified HRI was detected by αFLAG Western blotting. Cells were treated with proteasome inhibitor (CFZ, 2 μM) for 6 h prior to harvesting. Similar results in n = 2 independent experiments. d. The SIFI complex modifies HRI with ubiquitin chains predominantly linked to K48 of ubiquitin. Ubiquitylation of 35S-labeled HRI1–138-SUMO was analyzed as described above, but in the presence of indicated ubiquitin mutants (K0: all Lys residues mutated to Arg; K6only: all Lys residues except for K6 mutated to Arg). Experiment performed once. e. HRI and cDELE1 are degraded through the proteasome. Cells were analyzed for levels of stability reporters encoding HRI-GFP or cDELE1-GFP by flow cytometry. The proteasome inhibitor carfilzomib (2 μM) or the lysosome inhibitor bafilomycin A (700 nM) were added for 6 h as indicated. Similar results in n = 2 independent experiments. For gel source data, see Supplementary Fig. 1.