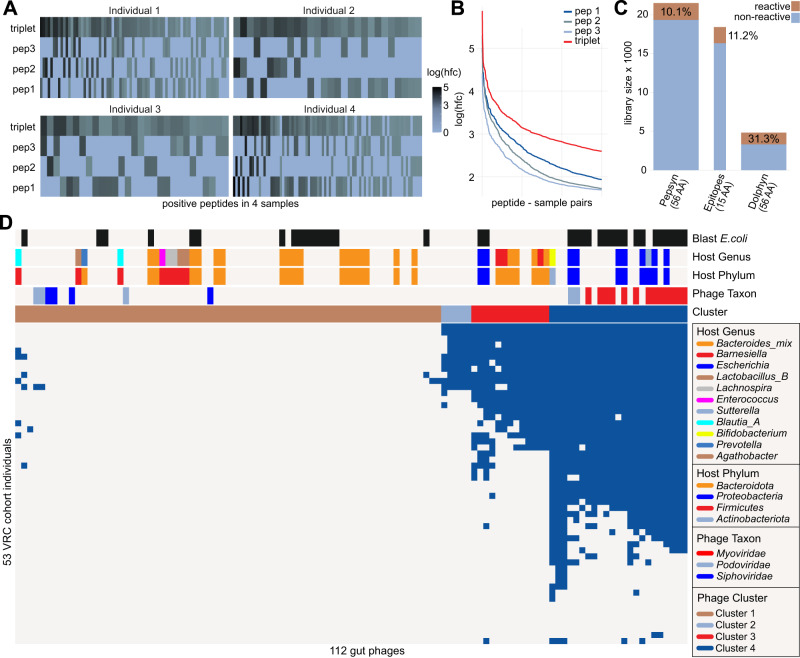
Fig. 4. Healthy individuals’ antibody reactivity to 112 gut phages in pilot library.
A Relationship between reactivities of stitched Dolphyn peptides and unstitched predicted epitope peptides. For four samples, all peptide quartets (three 15-mers and their stitched version) are shown where two or more peptides show reactivity. Peptide sets are ordered by highest to lowest reactivity from left to right. B The 15-mer peptides were stitched in the order of their probability score on the combined peptide, starting with the highest at position one. Accordingly, a difference in mean log reactivity score can be observed. Peptides are ordered by reactivity score per sample on the x-axis. C The phage proteomes in the pilot library were represented using three different approaches: Pepsyn, predicted epitopes, and Dolphyn. The Pepsyn library with regularly spaced 56-mer probes and is the largest but least reactive per peptide. The predicted epitopes alone achieve a similar proportion of reactive peptides despite the probes being small (15-mer amino acids (AA), depicted by width of bar). The Dolphyn library achieves a greater proportion of reactive probes than the alternatives. D Anti-phage antibody reactivities in healthy individuals of the Vaccine Research Center (VRC) cohort, depicted as binarized PhARscores, a phage-level aggregate reactivity score. The annotation on the top shows predicted phage properties and phage clusters. | Source data are provided as a Source Data file.