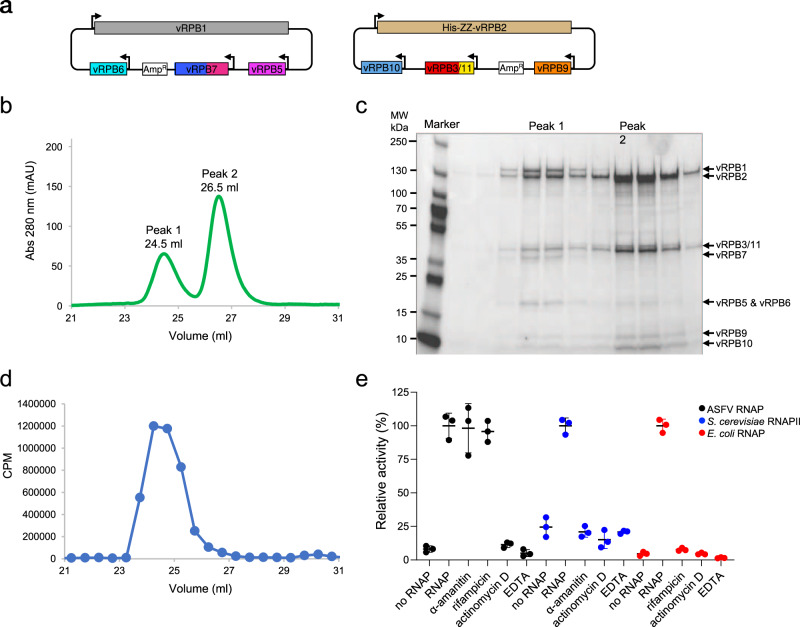
Fig. 1. Expression, purification, and biochemical characterisation of the ASFV RNAP.
a Overview of cloning strategy for the recombinant ASFV RNAP. The first bacmid-generating plasmid includes vRPB1 (grey), vRPB5 (magenta), vRPB6 (cyan), and vRPB7 (blue) with the CTD in deep pink. The second plasmid contains vRPB2 (tan), vRPB3-11 fusion (red and yellow, respectively), vRPB10 (cornflower blue) and vRPB9 (orange). vRPB2 includes an N-terminal cleavable ZZ-affinity tag. b UV profile of SEC purification. c SDS-PAGE analysis of SEC purification step reveals that the two peaks contain the complete RNAP (peak 1) and a subassembly (peak 2) lacking vRPB1, 5, 6 and 7, respectively. This is a representative gel, which was repeated six times. Source data are provided as a Source Data file. d Nonspecific in vitro transcription assay shows that only SEC peak 1 fractions contain robust transcription activity. Activity is reported as CPM (counts per minute) of radiolabelled [α−32P]-UTP incorporation as a function of the SEC elution volume (from panel b). The experiment was carried out only for the first purification. e Effect of RNAP inhibitors on ASFV core RNAP. Alpha-amanitin and rifampicin, both at 100 µM, do not inhibit the ASFV RNAP, while inhibition was observed for the DNA intercalator Actinomycin D (100 µM) and magnesium chelator EDTA (50 mM). Control experiments using S. cerevisiae RNAPII and E. coli RNAP were used to confirm alpha-amanitin and rifampicin inhibition. For all polymerases a negative control with only buffer (no RNAP lane) was prepared. Transcription activity is shown as % of radiolabelled [α−32P]-UTP incorporation relative to each RNA polymerase in the absence of inhibitors (labelled as RNAP) versus the same reaction without enzyme (no RNAP). Results were produced in triplicates and reported as the mean with corresponding standard deviation. Source data are provided as a Source Data file.