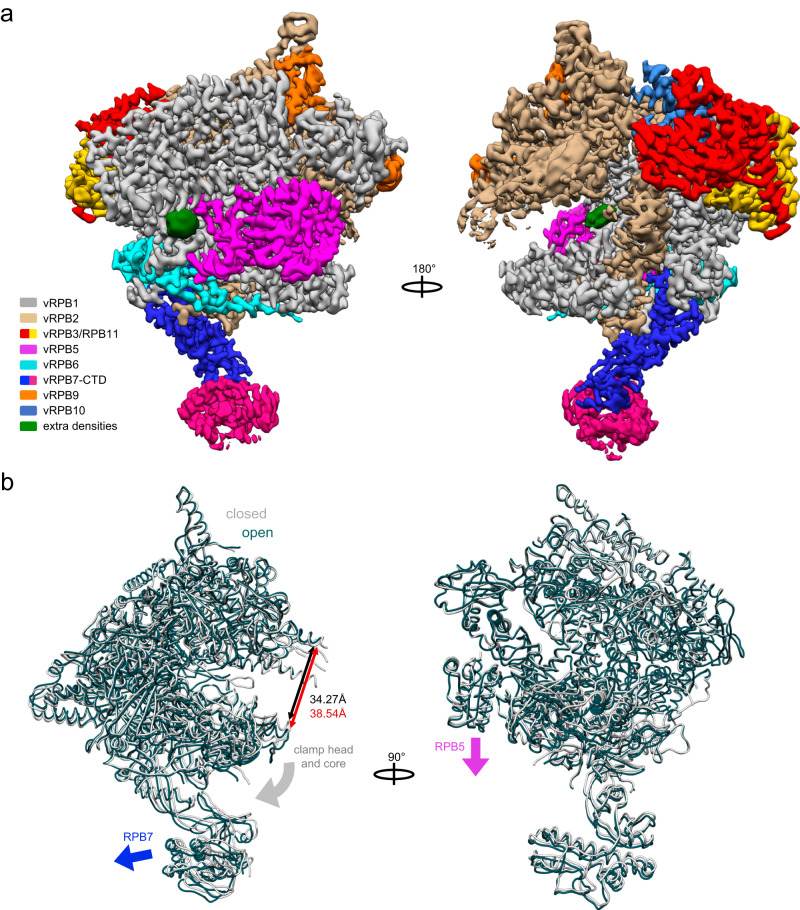
Fig. 2. The cryo-EM structures of the ASFV core RNA polymerase.
a The cryo-EM map of the 8-subunit RNAP with the subunits colour coded according to the legend. The EM map reveals two unidentified densities/ligands, shown in green. b Superposition of the structures corresponding to the closed and open conformations of the RNAP (grey and teal, respectively). Conformational changes involving movements of vRPB5 (magenta block arrow), vRPB7 (blue block arrow), as well as clamp head and core (grey block arrow) lead to the widening of the DNA-binding channel by 4.3 Å. The two conformations are shown in worm style and superimposed on the vRPB2 subunit. The width of the DNA-binding channel was measured between residues vRPB2 Val357 and vRPB1 Leu254 in Chimera v1.16.083.