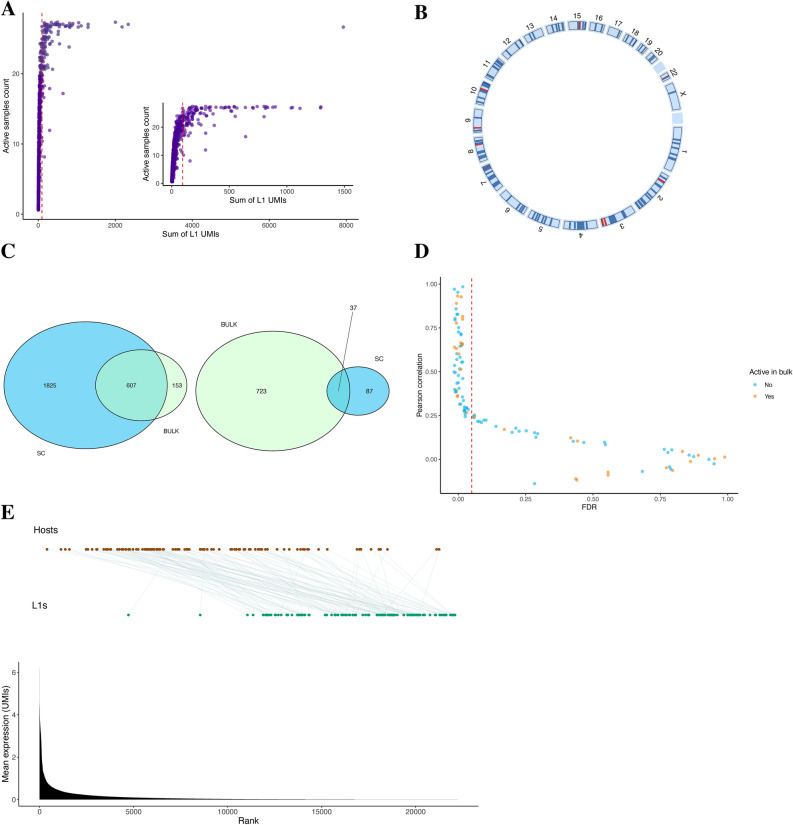
Figure 1.
Limited overlap of transcriptionally active L1 loci detected by scRNA-seq versus bulk RNA -seq. (A) The relationship between total number of L1 unique molecular identifiers (UMIs) and L1-expressing samples. Each dot corresponds to a single L1 locus. Dashed red line indicates the 95th percentile of the most active L1s. (B) Genomic locations of the 124 most highly active L1s. L1s with a total UMI count above 1000 are indicated with red, otherwise they are colored with blue. (C) Overlap between total (left panel) or top 5% expressed L1 loci (right panel) detected in scRNA-seq and bulk RNA-seq. (D. Correlation between expression of L1s and their overlapping genes based on scRNA-seq data. Dashed red line indicates False discovery rate (FDR) = 0.05. L1s detected in both single cell and bulk RNA-seq are marked in blue, whereas scRNA-seq specific L1s are marked in orange. (E) Mean expression of L1s and their overlapping genes based on single cell data. The statistics are based on all samples and cell types combined. Barplot shows ranked mean expression of all genes. Green dots denote L1s, whereas brown dots denote L1-overlapping genes, ordered by rank.