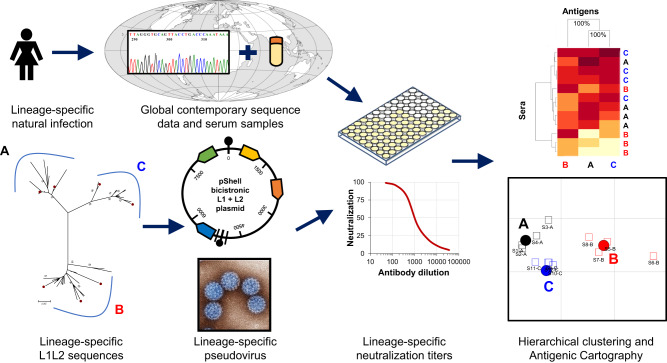
Fig. 1. Schematic depicting study execution.
Residual archived cervical samples from self-reported females collected for natural history or baseline vaccine studies that had previously been genotyped were assigned a specific lineage following whole genome or Sanger sequencing. Where available, contemporaneously collected serum (or plasma) samples representing defined lineages for each of the following genotypes (HPV16, 18, 31, 33, 45, 52, and 58) were assembled. Lineage-specific L1 and L2 sequences were downloaded from the NCBI database and bicistronic psheLL vectors containing the majority of L1 and L2 inserts together with a luciferase reporter plasmid was used to create lineage-specific PsVs following transfection of 293TT cells. Lineage-specific serum (or plasma) and lineage-specific PsV were used in neutralization assays to generate lineage-specific neutralization titers. Neutralizing antibody data were subjected to hierarchical clustering and antigenic cartography methods to create relational antigenic profiles for each genotype. World map courtesy of Tentotwo (https://commons.wikimedia.org/w/index.php?curid=21507855).