Abstract
We sought to study the binding constraints placed on the nine-zinc-finger protein transcription factor IIIA (TFIIIA) by a histone octamer. To this end, five overlapping fragments of the Xenopus laevis oocyte and somatic 5S rRNA genes were reconstituted into nucleosomes, and it was subsequently shown that nucleosome translational positioning is a major determinant of the binding of TFIIIA to the 5S rRNA genes. Furthermore, it was found that histone acetylation cannot override the TFIIIA binding constraints imposed by unfavorable translational positions.
Xenopus laevis produces two major types of 5S rRNA: the somatic type is synthesized in most cell types, whereas the oocyte type is synthesized during early oogenesis, during embryogenesis, and in certain tissue culture cell lines (14, 47). Each 5S rRNA type is transcribed from a distinct multigene family, and considerable research has focused on understanding the differential expression of these genes. One hypothesis suggests that the transcription complexes which form on the oocyte genes are relatively unstable compared to those of the somatic counterparts. As a result of this, the oocyte genes are transcribed only when transcription factor IIIA (TFIIIA) levels are relatively high, as is the case during oogenesis. In somatic cells, in which TFIIIA levels are much lower, the transcription complexes dissociate from the oocyte gene, allowing the subsequent assembly of repressive nucleosome structures which preclude further factor binding (47). Conflicting with this hypothesis are results which show that, at least in vitro, the RNA polymerase III transcription complexes, once formed, have similar stabilities on both the oocyte and somatic genes (35). Thus, at this time, the reasons for the differential transcription of these two gene families in X. laevis are not fully understood.
The results of 5S rRNA gene transcription studies, performed with both native and reconstituted chromatin as templates, have been conflicting. In one such study it was demonstrated that chromatin isolated from a Xenopus kidney cell line can serve as a template for transcription of the oocyte gene after the removal of histone H1 (34). In addition, it has been shown that incorporation of a somatic histone H1 variant into chromatin during embryogenesis results in specific repression of TFIIIA-activated oocyte 5S rRNA transcription (8). Furthermore, histone H1 has been shown to specifically repress transcription of oocyte genes in reconstituted chromatin while leaving the corresponding somatic genes unaffected (40). These results suggest that it is the presence of histone H1 which is responsible for the repression of oocyte transcription. This is not surprising considering that histone H1 is thought of as a repressor of transcription, although the exact mechanism of this repression is not known. In contrast to the above-mentioned work, studies with chromatin reconstituted in the absence of linker histones have shown that the presence of nucleosome core particles alone is sufficient to repress oocyte 5S rRNA gene transcription (18, 37). An explanation for these differing results is that it is the translational position of the histone octamers on the 5S rRNA gene which determines whether an active transcription complex can assemble and that repression by histone H1 is due to maintenance of a repressive translational position. One study using Xenopus borealis somatic 5S rRNA gene fragments reconstituted into dinucleosomes already supports this hypothesis (42). Further support is a study which mapped the positions of nucleosomes on the oocyte gene in both Xenopus nuclei and reconstituted chromatin. The results showed that the nucleosome translational position differs slightly depending on whether the oocyte genes are active (17).
The binding of TFIIIA to nucleosomally arranged DNA has been reported in the past in at least two instances (22, 32), but the results are conflicting. In one instance, a portion of the 5S promoter, containing nucleotides critical for the binding of TFIIIA, was located outside the 146-bp DNA fragment of the nucleosome core particle (32). In the other instance, a histone acetylation-mediated binding of TFIIIA to internal regions of the 146-bp core DNA was reported (22). It is the aim of this work to determine whether, in vitro, nucleosome translational position affects TFIIIA binding to the X. laevis 5S rRNA genes, thus supporting a model in which the differential translational position of nucleosomes on the oocyte and somatic 5S rRNA genes contributes to the differential regulation of these two gene families.
MATERIALS AND METHODS
Nucleosome reconstitution.
The salt gradient dialysis method (38) and the exchange method were the two nucleosome reconstitution techniques used in this study. The salt gradient dialysis method, which was used to reconstitute full-length 5S rRNA genes, used a DNA concentration of 100 μg/ml and a molar ratio of histone octamers to 5S rRNA genes of 3 for the oocyte gene and 4 for the somatic gene. The 720-bp oocyte and 880-bp somatic full-length 5S rRNA genes were isolated from HindIII digests of plasmids pXlo8 and pXlsII (28), respectively. For the exchange reaction, which was used to reconstitute the ∼200-bp 5S rRNA gene fragments, approximately 200 fmol of labeled DNA and 3 μg of cold nucleosome cores (isolated from either chicken erythrocytes, non-butyrate-treated HeLa cells, or butyrate-treated HeLa cells) were incubated in 25 μl of 0.8 M NaCl–50 mM Tris (pH 8)–1 mM β-mercaptoethanol–0.1 mM phenylmethylsulfonyl fluoride for 30 min at 37°C. The nucleosomes were then incubated at 4°C for 16 h, followed by stepwise dilution to 0.6 and 0.1 M NaCl by addition of 50 mM Tris (pH 8)–0.1 mM phenylmethylsulfonyl fluoride after 30-min intervals at 4°C.
In vitro transcription of reconstituted 5S rRNA genes.
HeLa cell nuclear in vitro transcription extracts were provided by Promega, and transcriptions were performed as per manufacturer’s instructions with 250 ng of template (either reconstituted nucleosomes or uncomplexed DNA). An MgCl2 concentration of 2 mM was used, and extracts were supplemented with 150 nM recombinant Xenopus TFIIIA. For each transcription reaction an internal control of 50 ng of cytomegalovirus (CMV) DNA was included.
Preparation and labeling of 5S rRNA gene fragments.
The five ∼200-bp, overlapping fragments of the X. laevis somatic and oocyte 5S rRNA genes were derived from plasmids pXlsII and pXlo(Δ3′+176) (6). Plasmids containing the oocyte 5S rRNA gene fragments designated Xlo(−83→+136) and Xlo(−38→+149) were created by exonuclease III digestion of pXlo(Δ3′+176). These exonuclease III digestions were performed independently on the 5′ and 3′ termini as described by others (33). Plasmids containing the X. laevis somatic 5S rRNA gene fragments designated Xls(−51→+147) and Xls(−1→+204) were created by ligation of pXlsII BanII/DdeI and EaeI/AluI restriction fragments, respectively, into the EcoRV site of Bluescript. A third plasmid containing the somatic 5S rRNA gene fragment designated pXls(−74→+147) was created by PCR amplification of a gene fragment from pXlsII with the New England Biolabs M13 Reverse Sequencing Primer (−24) and the oligonucleotide 5′CTTGGGAATTCAGCCCTGC3′. Following PCR, the amplified product was EcoRI/DdeI digested and ligated into Bluescript. As a result of these subcloning steps, each 5S rRNA gene fragment was flanked by an EcoRI site on the 5′ end and a HindIII site on the 3′ end of the gene. A 214-bp EcoRI/DdeI fragment derived from the plasmid pXP-10 (46) was used for the experiments involving the X. borealis 5S rRNA gene. This fragment can be described as Xbs(−75→+147) in our nomenclature.
Two techniques were employed to radioactively label the 5S rRNA gene fragments for this study. For the micrococcal nuclease digestions, for which internally labeled DNA was required, plasmid DNA was alkaline denatured and annealed to an M13 Sequencing Primer (−20). The subsequent labeling was performed in 20 μl of 1× restriction buffer with 1 mM dCTP–1 mM dGTP–1 mM dTTP–10 μM dATP–50 nM [α-32P]dATP–4 U of Klenow fragment, with incubation at 20°C for 30 min. The Klenow fragment was heat inactivated, and the labeled DNA was digested with EcoRI and HindIII. The DNA was electrophoretically purified on a 4% nondenaturing polyacrylamide gel and eluted from the gel slice by rotation for 16 h in 300 μl of 0.6 M ammonium acetate–0.1% sodium dodecyl sulfate–1 mM EDTA. By this technique a DNA fragment internally labeled on one strand was produced, which facilitated the interpretation of the results of the nucleosome translational position analysis. For DNase I footprinting analysis, DNA was 3′ end labeled at the EcoRI site for footprinting of the noncoding strand and at the HindIII site for footprinting of the coding strand.
Purification of TFIIIA and nucleosome core particles.
The purification of recombinant TFIIIA from Escherichia coli cells harboring the expression plasmid pTF3 was carried out as described previously (44). The technique of Ausió et al. (2) was used for the isolation of nucleosome core particles from chicken erythrocytes. Nucleosome core particles with low or high levels of acetylation were obtained from HeLa cells grown in the absence or presence of sodium butyrate as described by Ausió and Van Holde (4). The level of histone acetylation was analyzed by electrophoreses on a Triton-urea-acetic acid gel (7).
Determination of nucleosome translational position.
Nucleosome core particles reconstituted on the 5S rRNA gene fragments were adjusted to 1 mM CaCl2 and digested with micrococcal nuclease. The time of digestion and the amount of micrococcal nuclease were established from a previously determined time course of a digestion analysis carried out under the same conditions. Digestion was stopped and the DNA was deproteinized by adjusting the solution to 5 mM EDTA–0.25% sodium dodecyl sulfate and phenol-chloroform extracting. The approximately 146-bp micrococcal nuclease-resistant DNA fragments were electrophoretically purified on a 6% nondenaturing polyacrylamide gel and, after elution from the acrylamide matrix, precipitated and restriction enzyme digested. The digested fragments were phenol-chloroform extracted, ethanol precipitated, and resolved on an 8% acrylamide gel (acrylamide-bisacrylamide, 19:1) containing 8.3 M urea and 1× TBE (90 mM Tris-borate, 2 mM EDTA).
TFIIIA electrophoretic mobility shift assays.
Approximately 1-fmol amounts of labeled reconstituted nucleosomes were incubated in 10 μl of 20 mM Tris (pH 7.5)–70 mM NaCl–10 μM ZnCl2–6% glycerol–0.1 mg of bovine serum albumin per ml–2.5 mM dithiothreitol–0.07% Nonidet P-40–40 ng of poly(dI-dC) · poly(dI-dC) per μl for 20 min at room temperature with increasing amounts of TFIIIA. The binding reaction mixtures were loaded on a 0.75% agarose gel containing 0.5× TB (45 mM Tris-borate), and reactions were run at 3.5 V/cm at 20°C. The gels were dried at 50°C and autoradiographed. TFIIIA shifts of uncomplexed DNA were carried out in a similar manner with 100 ng of poly(dI-dC) · poly(dI-dC) per μl added to the binding reaction mixtures.
DNase I footprinting analysis.
Ten femtomoles of reconstituted nucleosomes was incubated with a 50-fold molar excess of TFIIIA in 20 μl of 20 mM Tris (pH 7.5)–70 mM NaCl–10 μM ZnCl2–6% glycerol–0.1 mg of bovine serum albumin per ml–2.5 mM dithiothreitol–0.07% Nonidet P-40 for 20 min at room temperature. Immediately thereafter, 1 μl of 1-ng/μl DNase I was added, and digestion was allowed to proceed for 1 min at room temperature. The reaction mixtures were immediately loaded onto a 4% nondenaturing gel, and reactions were run at 10 V/cm at room temperature. The gel was autoradiographed wet, and bands corresponding to untreated and TFIIIA-shifted nucleosomes were excised. The digested DNA was eluted as described earlier, ethanol precipitated, and resolved on an 8% acrylamide gel (acrylamide-bisacrylamide, 19:1) with 8.3 M urea and 1× TBE. Naked DNA, digested as described above, was used as a control, but in this case the gel purification step was omitted. Instead, the DNase I digests were heat inactivated at 90°C for 5 min, and the resulting DNA fragments were extracted with phenol-chloroform and ethanol precipitated. Maxam-Gilbert reactions of uncomplexed DNA were performed (33) for use as markers.
RESULTS
Differential transcription of oocyte and somatic 5S rRNA genes after nucleosome reconstitution.
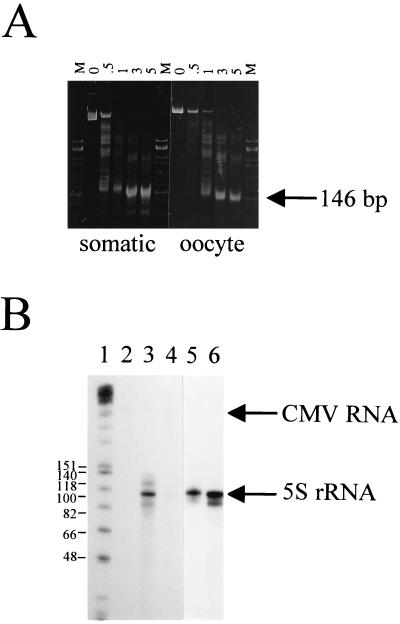
Previously it was shown that transcription of reconstituted 5S rRNA genes can occur only if, prior to reconstitution, a transcription complex or, minimally, TFIIIA is allowed to assemble on the intragenic promoter (18, 37). This suggests that, once bound to the DNA, TFIIIA prevents repressive nucleosome structures from forming on the DNA and argues against the idea that transcription initiation can be regulated by nucleosome translational position. To demonstrate that this is not the case, nucleosomes were reconstituted onto full-length 5S rRNA genes (880 and 720 bp in length for the somatic and oocyte genes, respectively) by a salt gradient dialysis method, and these genes were transcribed in HeLa cell nuclear extracts supplemented with TFIIIA. Figure 1A shows a micrococcal nuclease time course digestion which demonstrates that nucleosomes were present on the genes, as is indicated by 146-bp micrococcal nuclease resistant fragments. Results of transcription studies (Fig. 1B) indicate that nucleosomes reconstituted on the oocyte gene fragment repressed transcription of the 5S rRNA gene whereas those on the somatic gene fragment did not (compare lanes 4 and 6). Although it appears from this data that the differential transcription of these genes has been recreated in vitro, it must be noted that, because the nucleosomes were reconstituted with single copies of the 5S rRNA genes rather than the tandem units found in the cell, this is not necessarily an accurate representation of what is present in vivo. The results do suggest that in vitro reconstitution of nucleosomes on the 5S rRNA genes can result in both transcription-permissive and transcription-repressive chromatin structures.
FIG. 1.
Effect of nucleosome reconstitution on 5S rRNA gene transcription. (A) Micrococcal nuclease digestion of nucleosomes reconstituted on full-length X. laevis oocyte and somatic genes. Digestions were carried out at a nucleosome concentration of 0.1 mg/ml (DNA weight) and an enzyme concentration of 10 U/ml for the times (minutes) indicated above each lane. The resulting DNA fragments were deproteinized and electrophoresed on a 4% nondenaturing gel. Lanes M, HhaI-cut pBR322. (B) Approximately 250 ng of the oocyte (lanes 3 and 4) or somatic (lanes 5 and 6) 5S rRNA genes, either uncomplexed (lanes 3 and 5) or reconstituted with histones isolated from chicken erythrocytes (lanes 4 and 6), was transcribed in HeLa cell nuclear extracts supplemented with 150 nM recombinant Xenopus TFIIIA and 2 mM MgCl2. Each reaction mixture contained 50 ng of CMV DNA as an internal transcription control, which is visible only after longer exposures. Transcripts were analyzed by denaturing polyacrylamide gel electrophoresis (8% acrylamide and 8.3 M urea in 1× TBE). Lane 1, Klenow fragment-end-labeled HinfI-cut φX174 DNA (sizes of marker fragments are shown as numbers of nucleotides); lane 2, transcribed CMV DNA alone.
Determination of the translational position of reconstituted nucleosomes.
To determine whether nucleosome translational position can mediate TFIIIA binding, it was necessary to create several mononucleosome particles in which the position of the nucleosome relative to the intragenic promoter was varied. In previous investigations designed to study the effect of nucleosome position, synthetic DNA-bending sequences were used to position trans-acting factor binding sites at different locations with respect to the histone octamer (5, 24, 25, 39). Due to the fact that the TFIIIA binding site is relatively large, it would be difficult to introduce the site into a DNA sequence designed to phase a nucleosome without altering the nucleosome position. For this reason, the positions of nucleosomes on the 5S rRNA genes were varied instead by altering the fragments used for nucleosome reconstitution. In addition, fragments of both the oocyte and somatic 5S rRNA genes were used, as the two genes position nucleosomes differently due to their differing 5′ and 3′ flanking sequences and yet bind TFIIIA with similar affinities.
Previous to this work, the positions of nucleosomes on reconstituted Xenopus laevis oocyte gene fragments had not been published, and those of the somatic gene had been studied extensively albeit with conflicting results. According to Gottesfeld (16), mononucleosomes reconstituted onto different fragments of the somatic 5S rRNA gene occupy a region spanning nucleotides +20 to +200 with respect to the transcriptional start site. In contrast, when Lee et al. (22) reconstituted a nucleosome on a different X. laevis 5S rRNA gene fragment, they described the nucleosome position as further upstream, with a dyad axis at nucleotide +32. This suggests that the translational position of a mononucleosome on this gene is dependent on the DNA fragment chosen for the reconstitution. The gene fragments used in this study were Xlo(−83→+136), Xlo(−38→+149), Xls(−74→+147), Xls(−51→+147), and Xls(−1→+204), with the prefix indicating the source of the gene (Xlo referring to oocyte and Xls referring to somatic) and numbers representing the 5′ and 3′ ends of the DNA fragments in relation to the site of transcription initiation.
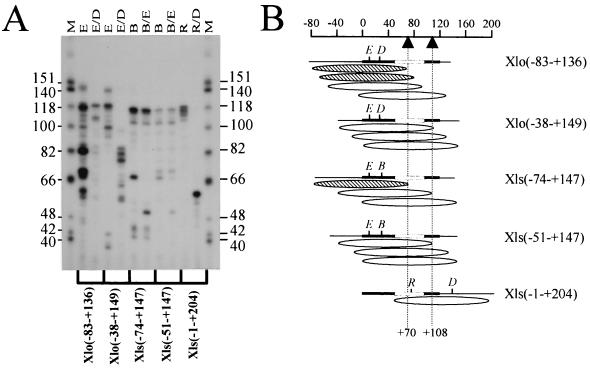
By using an approach similar to that of Dong et al. (13), the positions of mononucleosomes reconstituted by the exchange method on the 5S rRNA gene fragments were determined. Briefly, this technique involved digesting nucleosomes with micrococcal nuclease and mapping the position of the micrococcal nuclease-resistant fragment by digestion with one or more restriction enzymes. Figure 2A shows an example of the electrophoresis patterns obtained. After analysis of the restriction digestion patterns in several trials, the most abundant nucleosome positions on each fragment were determined (Fig. 2B). It is important to note that these positions are intrinsic to the DNA fragments used and are not meant to represent the positions that nucleosomes occupy in vivo.
FIG. 2.
Determination of nucleosome translational position on reconstituted X. laevis 5S rRNA gene fragments. (A) The region of DNA in direct association with the histone octamer was determined by digestion of an approximately 146-bp, internally labeled, micrococcal nuclease-resistant fragment with different combinations of restriction enzymes as indicated. The resulting restriction fragments were resolved by denaturing polyacrylamide gel electrophoresis (8% acrylamide and 8.3 M urea in 1× TBE). Lanes M, HinfI-cut φX174 DNA used as a marker (sizes of marker fragments are shown as numbers of nucleotides). The restriction enzymes used are indicated above the gel: E, EaeI; D, DdeI; B, Bsp1286; R, RsaI. (B) Schematic representation of the most predominant nucleosome positions on the five different fragments of the X. laevis oocyte and somatic 5S rRNA genes resulting from the electrophoretic analysis shown in panel A. Due to variations in base composition and thus radioactive labeling, the intensities of the bands in relation to the dATP contents of the fragments were considered in these calculations. The ellipsoids indicate the most abundant positions of the approximately 146-bp micrococcal nuclease-resistant fragments. The thick black lines represent the 5S rRNA coding sequence, and the open boxes indicate the intragenic TFIIIA binding site. Nucleotide positions relative to the transcription start site are indicated on the scale at the top, and the dashed vertical lines indicate positions +70 and +108. The hatched ellipsoids are those nucleosome positions postulated to allow TFIIIA binding, whereas open ellipsoids are those which are thought to be repressive to TFIIIA binding based on the results of You et al. (48).
Binding of TFIIIA to X. laevis 5S rRNA gene fragments after nucleosome reconstitutions.
Previous work reported that TFIIIA cannot bind the X. laevis somatic 5S rRNA gene after reconstitution with a complete histone octamer (16, 22). When a similar experiment was performed with the X. borealis somatic 5S rRNA gene, which has been shown to position a nucleosome differently from that of X. laevis (16, 32), the results were conflicting. Rhodes (32) was able to demonstrate TFIIIA binding to nucleosomal X. borealis somatic 5S rRNA genes, while Hayes and Wolffe (20) found the TFIIIA binding site to be blocked, after nucleosome reconstitution. Lee et al. (22) instead found that reconstitution with histone tetramers rather than complete octamers permits TFIIIA binding to the X. borealis gene but not the X. laevis gene. This difference was attributed to the differential translational positions of mononucleosomes on the two somatic genes, thus suggesting that nucleosome position can affect transcription factor binding. However, it was shown that TFIIIA can bind to both the X. laevis and X. borealis 5S rRNA genes after reconstitution with acetylated histone octamers (22). This suggests that the steric hindrance imposed by the nucleosome on TFIIIA binding to the X. laevis 5S rRNA gene can be overcome by acetylation.
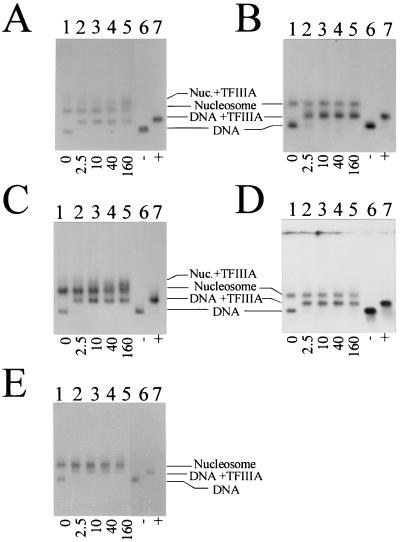
To test whether any of the nucleosomes reconstituted for this study could bind TFIIIA, electrophoretic mobility shift assays were performed (Fig. 3). The molar ratio of TFIIIA to 5S rRNA genes is as indicated at the bottom of the gels. It must be noted that the TFIIIA concentrations used in these studies are far less than what is present in vivo, since during early oogenesis, TFIIIA concentrations can reach 107 times the number of oocyte 5S rRNA genes (36), although much of this becomes complexed as the 7S particle. The higher concentrations used in this study were sufficient to completely shift the corresponding uncomplexed DNA alone (Fig. 3, lanes 6 and 7). Under these conditions, TFIIIA should saturate all of the available binding sites.
FIG. 3.
Analysis of the binding of TFIIIA to nucleosomes reconstituted onto the X. laevis 5S rRNA gene fragments by agarose gel electrophoresis. (A) Xlo(−83→+136); (B) Xlo(−38→+149); (C) Xls(−74→+147); (D) Xls(−51→+147); (E) Xls(−1→+204). Lanes 1 to 5, nucleosomes reconstituted with histones isolated from non-butyrate-treated HeLa cells in the absence (lane 1) or presence (lanes 2 to 5) of increasing amounts of TFIIIA (molar ratios are indicated below the gels). Lanes 6 and 7, corresponding DNA templates in the absence (−) or presence (+) of TFIIIA. The samples were incubated for 20 min at room temperature before being loaded on agarose gels.
Of the five gene fragments tested, only one oocyte [Xlo(−83→+136)] and one somatic [Xls(−74→+147)] gene fragment were able to bind TFIIIA after nucleosome reconstitution (Fig. 3A and C). However, in none of these cases could a complete shift of all nucleosomes be obtained, suggesting that either only a fraction of the nucleosomes are capable of binding TFIIIA or insufficient TFIIIA is present. The remaining 5S rRNA gene fragments, Xlo(−38→+149), Xls(−51→+147), and Xls(−1→+204), were unable to bind TFIIIA after nucleosome reconstitution even though in every instance the amount of transcription factor present was enough to completely shift the corresponding DNA alone (Fig. 3B, D, and E).
When reconstituted, the fragment Xlo(−83→+136) positioned mononucleosomes at four major locations (Fig. 2B) with 3′ boundaries at approximately positions +69, +80, +93, and +130 with respect to the transcriptional start site. TFIIIA bound to an uncomplexed X. borealis somatic 5S rRNA gene has been shown to occupy nucleotides +45 to +97 (32), with residues +81 to +91 forming the minimal requirements for TFIIIA binding (48). Thus, presumably, the fraction of mononucleosomes reconstituted on Xlo(−83→+136) which binds TFIIIA would be that with the TFIIIA binding site partially exposed (nucleosomes with 3′ boundaries at nucleotides +69 and +80). More evidence to support this conclusion is that fragment Xlo(−38→+149), which positions nucleosomes with boundaries at approximately nucleotides +110, +131, and +148, cannot bind TFIIIA. Of the three somatic gene fragments tested, only one, Xls(−74→+147), was able to bind TFIIIA after nucleosome reconstitution. This fragment positions a nucleosome predominantly at three sites with downstream boundaries at nucleotides +70, +108, and +146. The latter two positions, +108 and +146, were shared by nucleosomes reconstituted on Xls(−51→+147). However, this construct did not bind TFIIIA when existing in a nucleosomal form. This suggests that it is the nucleosome with the +70 downstream boundary on Xls(−74→+147) which permits TFIIIA binding.
Although from this data it is not possible to conclude exactly which nucleotides must be free in order for TFIIIA binding to occur, these results strongly suggest that nucleosomes positioned with their 3′ boundary upstream of position +70 are capable of binding TFIIIA whereas nucleosomes with their 3′ boundary downstream of position +108 cannot bind TFIIIA. This demonstrates that nucleosome translational positioning is a major determinant of the binding of TFIIIA to nucleosomal DNA.
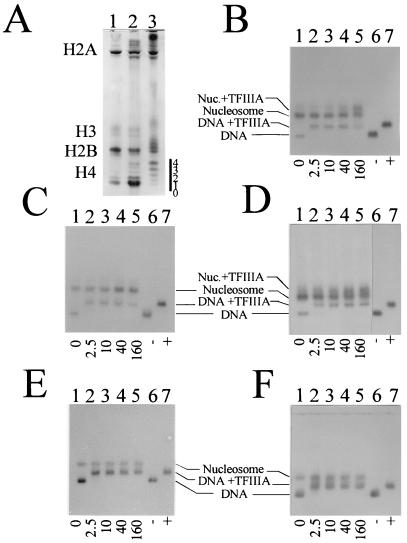
Lee et al. (22) demonstrated that blockage of TFIIIA binding by nucleosome reconstitution can be overcome by reconstitution with acetylated histones. Thus, in this study, the TFIIIA binding studies were repeated with the exception that histones from sodium butyrate-treated HeLa cells were used to determine whether histone acetylation could circumvent blockage of transcription factor binding. The histone composition and the level of histone acetylation are shown in Fig. 4A. As can be seen, the majority of histone H4 isolated from the sodium butyrate-treated cells existed in the tri- and tetra-acetylated forms. The results of electrophoretic mobility shift assays did not show any effect due to histone acetylation, as reconstitutions of nucleosomes with acetylated histones was unable to facilitate TFIIIA binding in the cases of Xlo(−38→+149), Xls(−51→+147), and Xls(−1→+204) (Fig. 4C, E, and F) or to enhance binding to Xlo(−83→+136) and Xls(−74→+147) (Fig. 4B and D).
FIG. 4.
(A) Acetic acid (6%)–urea (8 M)–Triton X-100 (8 mM) electrophoretic analysis of the histones from the nucleosome core particles used in the exchange reconstitutions. Lane 1, chicken erythrocyte core particles; lane 2, HeLa cell nucleosome core particles; lane 3, nucleosome core particles (fraction A [4]) from butyrate-treated HeLa cells. The number of acetyl groups on histone H4 is indicated by the numbers to the right of the gel. (B to F) Analysis of the binding of TFIIIA to nucleosomes containing hyperacetylated HeLa histones. The five X. laevis 5S rRNA gene fragments were tested for binding after reconstitution with nucleosome core particles isolated from sodium butyrate-treated HeLa cells. (B) Xlo(−83→+136); (C) Xlo(−38→+149); (D) Xls(−74→+147); (E) Xls(−51→+147); (F) Xls(−1→+204). Lanes 1, no TFIIIA; lanes 2 to 5, increasing amounts of TFIIIA (molar ratios are indicated below the gels); lanes 6 and 7, corresponding DNA templates in the absence (−) or presence (+) of TFIIIA. The samples were incubated for 20 min at room temperature before being loaded on agarose gels.
DNase I footprint analysis of the TFIIIA-5S rRNA gene-histone octamer ternary complex.
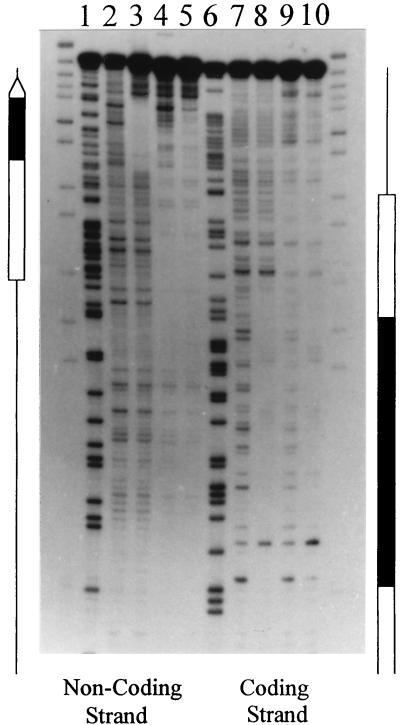
To rule out nonspecific binding in the mobility shift assays shown in Fig. 3 and 4, it was necessary to show correct contacts between TFIIIA and the DNA. To this end, DNase I footprinting analysis of the TFIIIA-nucleosome complex was performed with the oocyte 5S rRNA gene fragment Xlo(−83→+136). To ensure that this footprint represented the true TFIIIA-nucleosome complex, following nuclease digestion, the TFIIIA-nucleosome complex was purified by native gel electrophoresis. The DNase I digestion patterns of both the coding and noncoding strands are shown in Fig. 5. By comparing lanes 2 and 3 and lanes 7 and 8, the protection pattern of TFIIIA on the naked oocyte gene could be established. The transcription factor protected a region extending from nucleotide +45 to +91. Comparison between lanes 2 and 4 as well as lanes 7 and 9 in Fig. 5 shows an altered DNase I digestion pattern expected of overlapping nucleosome positions. The resolved digestion pattern of the TFIIIA-nucleosome complex seen in lanes 5 and 10 shows characteristics of both TFIIIA and nucleosome binding. The fact that there was TFIIIA-like protection over the entire TFIIIA binding site of the noncoding strand and not just nucleotides +81 to +91 indicates that all nine zinc fingers of TFIIIA were in contact with the DNA and not just fingers 1 to 3, which bind to nucleotides +81 to +91. This seems to suggest that TFIIIA unwrapped the DNA from the nucleosome to facilitate binding.
FIG. 5.
DNase I footprint analysis of the complexes formed by nucleosomes and/or TFIIIA on the Xlo(−83→+136) 5S rRNA gene fragment (Fig. 3B). Nucleosomes labeled at the 3′ end of the coding and noncoding strands were incubated in the presence of TFIIIA and subsequently digested with DNase I. The partially digested TFIIIA-nucleosome complexes were purified from free DNA and unbound nucleosomes by native gel electrophoresis (see Materials and Methods). The footprints for the coding and noncoding strands for naked DNA (lanes 2 and 7), TFIIIA-bound DNA (lanes 3 and 8), reconstituted nucleosome (lanes 4 and 9), and TFIIIA-nucleosome complexes (lanes 5 and 10) are shown. Also shown are the Maxam-Gilbert reactions of the labeled DNA (lanes 1 and 6). The 5S rRNA gene is indicated by the open arrow, and the TFIIIA binding site is indicated by a black box.
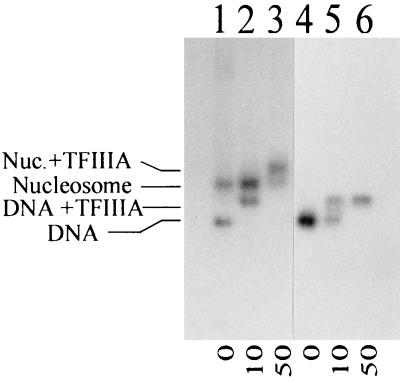
Binding of TFIIIA to X. borealis 5S rRNA gene fragment after nucleosome reconstitution.
The results of this study suggest that nucleosome translational position is a major determinant for the binding of TFIIIA to the 5S rRNA genes and that histone acetylation cannot override the TFIIIA binding constraints imposed by unfavorable translational positions. This is in direct conflict with the results of Lee et al. (22) that a nucleosome, positioned at approximately positions −70 to +79, was unable to bind TFIIIA if reconstituted with non-butyrate-treated HeLa cell histones. To test our hypothesis that TFIIIA can bind to nucleosomal 5S rRNA genes if the downstream boundary of the nucleosome is upstream of position +108, an electrophoretic mobility shift assay was performed with the same X. borealis gene fragment used by Lee et al. (22), reconstituted with histones from chicken erythrocytes. The results (Fig. 6) indicate that after nucleosome reconstitution, this gene fragment was almost completely shifted by TFIIIA. This is in agreement with our hypothesis that nucleosome translational position can modulate the binding of TFIIIA to nucleosomal 5S rRNA genes.
FIG. 6.
Analysis of the binding of TFIIIA to nucleosomes reconstituted onto the X. borealis 5S rRNA gene fragment by agarose gel electrophoresis, showing nucleosomes (lanes 1 to 3) or uncomplexed DNA (lanes 4 to 6) in the absence (lanes 1 and 4) or presence (lanes 2, 3, 5, and 6) of increasing amounts of TFIIIA. The nucleosomes used for the reconstitution were isolated from chicken erythrocytes, and the relative molar ratios of TFIIIA are indicated below the gel.
DISCUSSION
In order for transcription to be initiated, basal transcription factors and transcriptional trans activators must gain access to their cognate DNA sites. As a result of this, nucleosome position may be important for transcription initiation, and many studies have been performed to demonstrate the binding of trans-acting factors to sites within nucleosomal DNA (9, 21, 23–25, 27, 29, 39, 45). It has been shown that in the case of the glucocorticoid receptor, the translational position of the nucleosome is important for determining factor accessibility, as receptor binding to response elements near the dyad axis was less favorable than binding to other sites (25). In binding to DNA, the glucocorticoid receptor dimerizes, placing the DNA binding domains in two adjacent major grooves along one face of the DNA (26), and thus the association of a nucleosome with the other face does not necessarily preclude factor binding. The binding of TFIIIA to the 5S gene poses an additional problem. TFIIIA binds to a site which spans approximately five turns of DNA and consists of three elements: the A block (nucleotides +50 to +64), the IE element (+67 to +72), and the C block (+80 to +96). Current models for TFIIIA binding (10, 19) suggest that at least one complete turn of the DNA at each end of the intragenic promoter is bound on all faces of the helix by three zinc fingers in the major groove. Thus, unlike the case for the glucocorticoid receptor, complete binding of TFIIIA requires access to all sides of the DNA, and full binding would not seem possible if the entire TFIIIA binding site is associated with a nucleosome.
The main focus of this work was to investigate the effect of nucleosome translational position on TFIIIA binding. Each of the two oocyte fragments and three somatic fragments analyzed was tested for TFIIIA binding before and after nucleosome reconstitution. Our in vitro results demonstrated that in cases where the nucleosome was positioned in such a manner that the C block of the 5S rRNA gene intragenic promoter was within the 146 bp of DNA protected from micrococcal nuclease, TFIIIA could not bind the DNA (Fig. 2B and 3). A similar result was seen by Gottesfeld (16), who showed that a nucleosome with the translational position of nucleotides +20 to +200 on the X. laevis 5S rRNA gene blocks TFIIIA binding. Second, our work suggests that in those instances where at least the C block or more of the TFIIIA binding site was outside this region of DNA protected by the histone core, TFIIIA could bind (Fig. 2B and 3). This finding is in agreement with previous results (32) with the X. borealis 5S rRNA gene. DNase I footprinting (Fig. 5) showed that in one such instance both the IE element and the A block were bound by TFIIIA. This suggests that, upon binding to the C block, TFIIIA is able to unwind the DNA from the nucleosome to gain access to DNA sequences which are intimately associated with the histone octamer. This may occur by a mechanism of cooperative binding such as that already described by others (30, 31) for the binding of eukaryotic regulatory proteins to nucleosomal target sites.
The acetylation of core histones has long been known to be associated with transcriptionally active genes (for reviews, see references 1, 12, 41, and 43). Several recent experiments have shown that one of the roles of acetylation is to enhance the accessibility of DNA to transcription factors (22, 45). One such study used the TFIIIA-5S rRNA gene system as a model (22) and showed that in this case, reconstitution of the X. borealis and X. laevis somatic 5S rRNA genes with acetylated nucleosomes allows TFIIIA binding whereas reconstitution with nonacetylated nucleosomes prevents binding of this transcription factor. In our work, an attempt to reproduce these results found that in no case could histone acetylation overcome the blockage of TFIIIA binding after nucleosome reconstitution. Furthermore, it was shown in this study that TFIIIA was able to bind nucleosomal X. borealis genes in the absence of histone hyperacetylation. Although this supports the results of Rhodes (32), it is in direct conflict with those of Lee et al. (22). Differences between the experiments of Lee et al. (22) and our work include the use of high concentrations of MgCl2 and more dilute nucleosome concentrations by Lee et al. (22) (in our study an approximately 100-fold excess of nucleosome cores to labeled DNA was used for exchange reconstitutions, whereas Lee et al. [22] used only a 5-fold excess). Both of these factors have been shown to lead to disruption of histone-DNA contacts (3, 11, 15). An additional factor which could explain the differing results is the use of a slightly higher TFIIIA concentration in our study than in that of Lee et al. (22).
The results of this work support a model in which the differential translational positions of nucleosomes on the oocyte and somatic 5S rRNA genes could contribute to the differential regulation of these two gene families. Results already exist which strongly suggest that it is the preference of histone H1 for the oocyte gene which is responsible for the differential regulation of these genes (8, 34, 40), but the actual mechanism of H1-mediated repression is not known. Previous work has shown that histone H1-mediated reduction in nucleosome mobility is responsible for repression of transcription of a dinucleosome reconstituted onto a dimerized X. borealis somatic 5S rRNA gene (42). Reduced mobility would be expected to affect transcription only if certain translational positions restrict access of transcription factors to their cognate binding sites.
ACKNOWLEDGMENTS
We are very grateful to Nik Veldhoen and Paul Romaniuk for assistance in the preparation of TFIIIA and to Don Brown for the plasmids used in this work.
This work was supported by a grant from the Medical Research Council of Canada (MT-13104) to J.A.
REFERENCES
- 1.Ausió J. Structure and dynamics of transcriptionally active chromatin. J Cell Sci. 1992;102:1–5. doi: 10.1242/jcs.102.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Ausió J, Dong F, van Holde K E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J Mol Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausió J, Seger D S, Eisenberg H. Nucleosome core particle stability and conformational change. J Mol Biol. 1984;176:77–104. doi: 10.1016/0022-2836(84)90383-8. [DOI] [PubMed] [Google Scholar]
- 4.Ausió J, van Holde K E. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986;25:1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- 5.Blomquist P, Li Q, Wrange O. The affinity of nuclear factor 1 for its DNA site is drastically reduced by nucleosome organization irrespective of its rotational or translational position. J Biol Chem. 1996;271:153–159. doi: 10.1074/jbc.271.1.153. [DOI] [PubMed] [Google Scholar]
- 6.Bogenhagen D F, Brown D D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- 7.Bonner W M, West M H P, Stedman J D. Two dimensional gel analysis of histones in acid extracts of nuclei, cells and tissues. Eur J Biochem. 1980;109:17–23. doi: 10.1111/j.1432-1033.1980.tb04762.x. [DOI] [PubMed] [Google Scholar]
- 8.Bouvet P, Dimitrov S, Wolffe A P. Specific regulation of Xenopus chromosomal 5S rRNA gene transcription in vivo by histone H1. Genes Dev. 1994;8:1147–1159. doi: 10.1101/gad.8.10.1147. [DOI] [PubMed] [Google Scholar]
- 9.Chen H, Li N, Workman J L. A histone-binding protein, nucleoplasmin, stimulates transcription factor binding to nucleosomes and factor induced nucleosome disassembly. EMBO J. 1994;13:380–390. doi: 10.1002/j.1460-2075.1994.tb06272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens K R, Liao X, Wolf V, Wright P E, Gottesfeld J M. Definition of the binding site of individual zinc fingers in the transcription factor IIIA-5S RNA gene complex. Proc Natl Acad Sci USA. 1992;89:10822–10826. doi: 10.1073/pnas.89.22.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotton R W, Hamkalo B A. Nucleosome dissociation at physiological ionic strengths. Nucleic Acids Res. 1981;9:445–457. doi: 10.1093/nar/9.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Csordas A. On the biological role of histone acetylation. Biochem J. 1990;265:23–38. doi: 10.1042/bj2650023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong F, Hansen J C, van Holde K E. DNA and protein determinants of nucleosome positioning on sea urchin 5S rRNA gene sequences in vitro. Proc Natl Acad Sci USA. 1990;87:5724–5728. doi: 10.1073/pnas.87.15.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ford P J, Mathieson T. Control of 5S RNA synthesis in Xenopus laevis. Nature. 1976;261:433–435. doi: 10.1038/261433a0. [DOI] [PubMed] [Google Scholar]
- 15.Godde J S, Wolffe A P. Disruption of reconstituted nucleosomes. J Biol Chem. 1995;270:27399–27402. doi: 10.1074/jbc.270.46.27399. [DOI] [PubMed] [Google Scholar]
- 16.Gottesfeld J M. DNA sequence-directed nucleosome reconstitution on 5S RNA genes of Xenopus laevis. Mol Cell Biol. 1987;7:1612–1622. doi: 10.1128/mcb.7.5.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesfeld J M, Bloomer L S. Nonrandom alignment of nucleosomes on 5S RNA genes of X. laevis. Cell. 1980;21:751–760. doi: 10.1016/0092-8674(80)90438-9. [DOI] [PubMed] [Google Scholar]
- 18.Gottesfeld J M, Bloomer L S. Assembly of transcriptionally active 5S RNA gene chromatin in vitro. Cell. 1982;28:781–791. doi: 10.1016/0092-8674(82)90057-5. [DOI] [PubMed] [Google Scholar]
- 19.Hayes J J, Tullius T D. Structure of the TFIIIA-5S DNA complex. J Mol Biol. 1992;227:407–417. doi: 10.1016/0022-2836(92)90897-s. [DOI] [PubMed] [Google Scholar]
- 20.Hayes J J, Wolffe A P. Histones H2A/H2B inhibit the interaction of transcription factor IIIA with the Xenopus borealis somatic 5S RNA gene in a nucleosome. Proc Natl Acad Sci USA. 1992;89:1229–1233. doi: 10.1073/pnas.89.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juan L-J, Utley R T, Adams C C, Vettese-Dadey M, Workman J L. Differential repression of transcription factor binding by histone H1 is regulated by the core histone amino termini. EMBO J. 1994;13:6031–6040. doi: 10.1002/j.1460-2075.1994.tb06949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 23.Li B, Adams C C, Workman J L. Nucleosome binding by the constitutive transcription factor Sp1. J Biol Chem. 1994;269:7756–7763. [PubMed] [Google Scholar]
- 24.Li Q, Wrange O. Translational positioning of a nucleosomal glucocorticoid response element modulates glucocorticoid receptor affinity. Genes Dev. 1993;7:2471–2482. doi: 10.1101/gad.7.12a.2471. [DOI] [PubMed] [Google Scholar]
- 25.Li Q, Wrange O. The accessibility of a glucocorticoid response element dependent on its rotational positioning. Mol Cell Biol. 1995;15:4375–4384. doi: 10.1128/mcb.15.8.4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luisi B F, Xu W X, Otwinowski Z, Freedman L P, Yamamoto K R, Sigler P B. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 27.Perlmann T, Wrange O. Specific glucocorticoid receptor binding to DNA reconstituted in a nucleosome. EMBO J. 1988;7:3073–3079. doi: 10.1002/j.1460-2075.1988.tb03172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson R C, Doering J L, Brown D D. Characterization of two Xenopus somatic 5S DNAs and one minor oocyte-specific 5S DNA. Cell. 1980;20:131–141. doi: 10.1016/0092-8674(80)90241-x. [DOI] [PubMed] [Google Scholar]
- 29.Pina B, Bruggemeier U, Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- 30.Polach K J, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 31.Polach K J, Widom J. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J Mol Biol. 1996;258:800–812. doi: 10.1006/jmbi.1996.0288. [DOI] [PubMed] [Google Scholar]
- 32.Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985;4:3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Schlissel M S, Brown D D. The transcriptional regulation of Xenopus 5S RNA genes in chromatin: the role of active stable transcription complexes and histone H1. Cell. 1984;37:903–913. doi: 10.1016/0092-8674(84)90425-2. [DOI] [PubMed] [Google Scholar]
- 35.Seidel C W, Peck L J. Kinetic control of 5S RNA gene transcription. J Mol Biol. 1992;227:1009–1018. doi: 10.1016/0022-2836(92)90517-n. [DOI] [PubMed] [Google Scholar]
- 36.Shastry B S, Honda B M, Roeder R G. Altered levels of a 5S gene-specific transcription factor, TFIIIA, during oogenesis and embryonic development of Xenopus laevis. J Biol Chem. 1984;259:11373–11382. [PubMed] [Google Scholar]
- 37.Stunkel W, Kober I, Kauer M, Taimor G, Seifart K H. Human TFIIIA alone is sufficient to prevent nucleosome repression of a homologous 5S gene. Nucleic Acids Res. 1995;23:109–116. doi: 10.1093/nar/23.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatchell K, van Holde K E. Reconstitution of chromatin core particles. Biochemistry. 1977;16:5295–5303. doi: 10.1021/bi00643a021. [DOI] [PubMed] [Google Scholar]
- 39.Taylor I C A, Workman J L, Schuetz T J, Kingston R E. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991;5:1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- 40.Tomaszewski R, Jermanowski A. The AT-rich flanks of the oocyte-type 5S RNA gene of Xenopus laevis act as a strong signal for histone H1-mediated chromatin reorganization in vitro. Nucleic Acids Res. 1997;25:458–465. doi: 10.1093/nar/25.3.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner B M. Histone acetylation and control of gene expression. J Cell Sci. 1991;99:13–20. doi: 10.1242/jcs.99.1.13. [DOI] [PubMed] [Google Scholar]
- 42.Ura K, Hayes J J, Wolffe A P. A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO J. 1995;14:3752–3765. doi: 10.1002/j.1460-2075.1995.tb00045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Holde K E. Chromatin. Berlin, Germany: Springer-Verlag; 1988. [Google Scholar]
- 44.Veldhoen N, You Q M, Setzer D R, Romaniuk P J. Contribution of individual base pairs to the interaction of TFIIIA with the Xenopus 5S RNA gene. Biochemistry. 1994;33:7568–7575. doi: 10.1021/bi00190a009. [DOI] [PubMed] [Google Scholar]
- 45.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J L. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 46.Wolffe A P, Jordan E, Brown D D. A bacteriophage RNA polymerase transcribes through a Xenopus 5S RNA gene transcription complex without disrupting it. Cell. 1986;44:381–389. doi: 10.1016/0092-8674(86)90459-9. [DOI] [PubMed] [Google Scholar]
- 47.Wormington W M, Brown D D. Onset of 5S RNA gene regulation during Xenopus embryogenesis. Dev Biol. 1983;99:248–257. doi: 10.1016/0012-1606(83)90273-7. [DOI] [PubMed] [Google Scholar]
- 48.You Q, Veldhoen N, Baudin F, Romaniuk P J. Mutations in 5S DNA and 5S RNA have different effects on the binding of Xenopus transcription factor IIIA. Biochemistry. 1991;30:2495–2500. doi: 10.1021/bi00223a028. [DOI] [PubMed] [Google Scholar]